The insertion Green Monster (iGM) method for expression of multiple exogenous genes in yeast
- PMID: 24776987
- PMCID: PMC4455768
- DOI: 10.1534/g3.114.010868
The insertion Green Monster (iGM) method for expression of multiple exogenous genes in yeast
Abstract
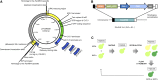
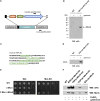
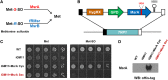
Being a simple eukaryotic organism, Saccharomyces cerevisiae provides numerous advantages for expression and functional characterization of proteins from higher eukaryotes, including humans. However, studies of complex exogenous pathways using yeast as a host have been hampered by the lack of tools to engineer strains expressing a large number of genetic components. In addition to inserting multiple genes, it is often desirable to knock out or replace multiple endogenous genes that might interfere with the processes studied. Here, we describe the "insertion Green Monster" (iGM) set of expression vectors that enable precise insertion of many heterologous genes into the yeast genome in a rapid and reproducible manner and permit simultaneous replacement of selected yeast genes. As a proof of principle, we have used the iGM method to replace components of the yeast pathway for methionine sulfoxide reduction with genes encoding the human selenoprotein biosynthesis machinery and generated a single yeast strain carrying 11 exogenous components of the selenoprotein biosynthetic pathway in precisely engineered loci.
Keywords: Saccharomyces cerevisiae; flow cytometry; green fluorescent protein; multi-gene insertions; synthetic biology.
Copyright © 2014 Labunskyy et al.
Figures

References
-
- Driscoll D. M., Copeland P. R., 2003. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23: 17–40. - PubMed
-
- Engels B., Dahm P., Jennewein S., 2008. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 10: 201–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
