Crystal structure of Mox-1, a unique plasmid-mediated class C β-lactamase with hydrolytic activity towards moxalactam
- PMID: 24777102
- PMCID: PMC4068568
- DOI: 10.1128/AAC.02363-13
Crystal structure of Mox-1, a unique plasmid-mediated class C β-lactamase with hydrolytic activity towards moxalactam
Abstract
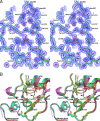
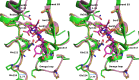
Mox-1 is a unique plasmid-mediated class C β-lactamase that hydrolyzes penicillins, cephalothin, and the expanded-spectrum cephalosporins cefepime and moxalactam. In order to understand the unique substrate profile of this enzyme, we determined the X-ray crystallographic structure of Mox-1 β-lactamase at a 1.5-Å resolution. The overall structure of Mox-1 β-lactamase resembles that of other AmpC enzymes, with some notable exceptions. First, comparison with other enzymes whose structures have been solved reveals significant differences in the composition of amino acids that make up the hydrogen-bonding network and the position of structural elements in the substrate-binding cavity. Second, the main-chain electron density is not observed in two regions, one containing amino acid residues 214 to 216 positioned in the Ω loop and the other in the N terminus of the B3 β-strand corresponding to amino acid residues 303 to 306. The last two observations suggest that there is significant structural flexibility of these regions, a property which may impact the recognition and binding of substrates in Mox-1. These important differences allow us to propose that the binding of moxalactam in Mox-1 is facilitated by the avoidance of steric clashes, indicating that a substrate-induced conformational change underlies the basis of the hydrolytic profile of Mox-1 β-lactamase.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

