Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation
- PMID: 24778178
- PMCID: PMC4067165
- DOI: 10.1074/jbc.M114.550350
Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation
Abstract
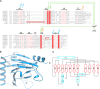
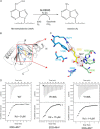
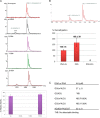
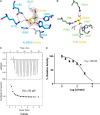
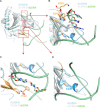
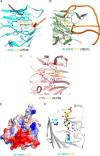
N(6)-Methyladenosine (m(6)A) is the most prevalent internal RNA modification in eukaryotes. ALKBH5 belongs to the AlkB family of dioxygenases and has been shown to specifically demethylate m(6)A in single-stranded RNA. Here we report crystal structures of ALKBH5 in the presence of either its cofactors or the ALKBH5 inhibitor citrate. Catalytic assays demonstrate that the ALKBH5 catalytic domain can demethylate both single-stranded RNA and single-stranded DNA. We identify the TCA cycle intermediate citrate as a modest inhibitor of ALKHB5 (IC50, ∼488 μm). The structural analysis reveals that a loop region of ALKBH5 is immobilized by a disulfide bond that apparently excludes the binding of dsDNA to ALKBH5. We identify the m(6)A binding pocket of ALKBH5 and the key residues involved in m(6)A recognition using mutagenesis and ITC binding experiments.
Keywords: Crystal Structure; Dioxygenase; Isothermal Titration Calorimetry (ITC); RNA Catalysis; RNA Methylation; Tricarboxylic Acid Cycle (TCA Cycle) (Krebs Cycle).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition.J Biol Chem. 2014 Apr 25;289(17):11571-11583. doi: 10.1074/jbc.M113.546168. Epub 2014 Mar 10. J Biol Chem. 2014. PMID: 24616105 Free PMC article.
-
Preparation of Human Nuclear RNA m⁶A Methyltransferases and Demethylases and Biochemical Characterization of Their Catalytic Activity.Methods Enzymol. 2015;560:117-30. doi: 10.1016/bs.mie.2015.03.013. Epub 2015 Apr 25. Methods Enzymol. 2015. PMID: 26253968 Free PMC article.
-
Crystal structure of the RNA demethylase ALKBH5 from zebrafish.FEBS Lett. 2014 Mar 18;588(6):892-8. doi: 10.1016/j.febslet.2014.02.021. Epub 2014 Feb 20. FEBS Lett. 2014. PMID: 24561204 Free PMC article.
-
RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential.J Hematol Oncol. 2022 Jan 21;15(1):8. doi: 10.1186/s13045-022-01224-4. J Hematol Oncol. 2022. PMID: 35063010 Free PMC article. Review.
-
The dynamic epitranscriptome: N6-methyladenosine and gene expression control.Nat Rev Mol Cell Biol. 2014 May;15(5):313-26. doi: 10.1038/nrm3785. Epub 2014 Apr 9. Nat Rev Mol Cell Biol. 2014. PMID: 24713629 Free PMC article. Review.
Cited by
-
Advances in the role of m6A RNA modification in cancer metabolic reprogramming.Cell Biosci. 2020 Oct 12;10:117. doi: 10.1186/s13578-020-00479-z. eCollection 2020. Cell Biosci. 2020. PMID: 33062255 Free PMC article. Review.
-
The m6A‑methylase complex and mRNA export.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):319-328. doi: 10.1016/j.bbagrm.2018.09.008. Epub 2018 Oct 2. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30290229 Free PMC article. Review.
-
Functions of RNA N6-methyladenosine modification in cancer progression.Mol Biol Rep. 2019 Apr;46(2):2567-2575. doi: 10.1007/s11033-019-04655-4. Epub 2019 Mar 25. Mol Biol Rep. 2019. PMID: 30911972 Review.
-
m6A RNA modification in transcription regulation.Transcription. 2021 Oct;12(5):266-276. doi: 10.1080/21541264.2022.2057177. Epub 2022 Apr 5. Transcription. 2021. PMID: 35380917 Free PMC article. Review.
-
m6A-Regulator Expression Signatures Identify a Subset of Follicular Lymphoma Harboring an Exhausted Tumor Microenvironment.Front Immunol. 2022 Jun 6;13:922471. doi: 10.3389/fimmu.2022.922471. eCollection 2022. Front Immunol. 2022. PMID: 35734168 Free PMC article.
References
-
- Globisch D., Pearson D., Hienzsch A., Brückl T., Wagner M., Thoma I., Thumbs P., Reiter V., Kneuttinger A. C., Müller M., Sieber S. A., Carell T. (2011) Systems-based analysis of modified tRNA bases. Angewandte Chemie 50, 9739–9742 - PubMed
-
- Iwanami Y., Brown G. M. (1968) Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 126, 8–15 - PubMed
-
- Saneyoshi M., Harada F., Nishimura S. (1969) Isolation and characterization of N6-methyladenosine from Escherichia coli valine transfer RNA. Biochim. Biophys. Acta 190, 264–273 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

