Assembly of the respiratory mucin MUC5B: a new model for a gel-forming mucin
- PMID: 24778189
- PMCID: PMC4047408
- DOI: 10.1074/jbc.M114.566679
Assembly of the respiratory mucin MUC5B: a new model for a gel-forming mucin
Abstract
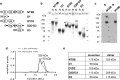
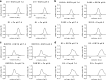
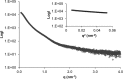
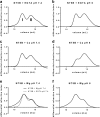
Mucins are essential components in mucus gels that form protective barriers at all epithelial surfaces, but much remains unknown about their assembly, intragranular organization, and post-secretion unfurling to form mucus. MUC5B is a major polymeric mucin expressed by respiratory epithelia, and we investigated the molecular mechanisms involved during its assembly. Studies of intact polymeric MUC5B revealed a single high affinity calcium-binding site, distinct from multiple low affinity sites on each MUC5B monomer. Self-diffusion studies with intact MUC5B showed that calcium binding at the protein site catalyzed reversible cross-links between MUC5B chains to form networks. The site of cross-linking was identified in the MUC5B D3-domain as it was specifically blocked by D3 peptide antibodies. Biophysical analysis and single particle EM of recombinant MUC5B N terminus (D1D2D'D3; NT5B) and subdomains (D1, D1-D2, D2-D'-D3, and D3) generated structural models of monomers and disulfide-linked dimers and suggested that MUC5B multimerizes by disulfide linkage between D3-domains to form linear polymer chains. Moreover, these analyses revealed reversible homotypic interactions of NT5B at low pH and in high calcium, between disulfide-linked NT5B dimers, but not monomers. These results enable a model of MUC5B to be derived, which predicts mechanisms of mucin intracellular assembly and storage, which may be common to the other major gel-forming polymeric mucins.
Keywords: Analytical Ultracentrifugation; Cystic Fibrosis; Goblet Cell; Mucin; Mucus; Recombinant Protein Expression; Single Particle Analysis.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Mucins: the frontline defence of the lung.Biochem Soc Trans. 2018 Oct 19;46(5):1099-1106. doi: 10.1042/BST20170402. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154090 Free PMC article. Review.
-
The MUC5B mucin polymer is dominated by repeating structural motifs and its topology is regulated by calcium and pH.Sci Rep. 2019 Nov 22;9(1):17350. doi: 10.1038/s41598-019-53768-0. Sci Rep. 2019. PMID: 31758042 Free PMC article.
-
The C-terminal dimerization domain of the respiratory mucin MUC5B functions in mucin stability and intracellular packaging before secretion.J Biol Chem. 2019 Nov 8;294(45):17105-17116. doi: 10.1074/jbc.RA119.010771. Epub 2019 Sep 30. J Biol Chem. 2019. PMID: 31570524 Free PMC article.
-
Biosynthesis of the polymeric gel-forming mucin MUC5B.Am J Physiol Lung Cell Mol Physiol. 2016 May 15;310(10):L993-L1002. doi: 10.1152/ajplung.00046.2016. Epub 2016 Mar 18. Am J Physiol Lung Cell Mol Physiol. 2016. PMID: 26993521 Free PMC article.
-
Respiratory tract mucins: structure and expression patterns.Novartis Found Symp. 2002;248:76-88; discussion 88-93, 277-82. Novartis Found Symp. 2002. PMID: 12568489 Review.
Cited by
-
New developments in goblet cell mucus secretion and function.Mucosal Immunol. 2015 Jul;8(4):712-9. doi: 10.1038/mi.2015.32. Epub 2015 Apr 15. Mucosal Immunol. 2015. PMID: 25872481 Free PMC article. Review.
-
Membrane mucins of the intestine at a glance.J Cell Sci. 2020 Mar 13;133(5):jcs240929. doi: 10.1242/jcs.240929. J Cell Sci. 2020. PMID: 32169835 Free PMC article. Review.
-
Emerging cell and molecular targets for treating mucus hypersecretion in asthma.Allergol Int. 2024 Jul;73(3):375-381. doi: 10.1016/j.alit.2024.04.002. Epub 2024 May 1. Allergol Int. 2024. PMID: 38692992 Free PMC article. Review.
-
Mucins: the frontline defence of the lung.Biochem Soc Trans. 2018 Oct 19;46(5):1099-1106. doi: 10.1042/BST20170402. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154090 Free PMC article. Review.
-
Hypercapnia modulates cAMP signalling and cystic fibrosis transmembrane conductance regulator-dependent anion and fluid secretion in airway epithelia.J Physiol. 2016 Mar 15;594(6):1643-61. doi: 10.1113/JP271309. Epub 2015 Dec 20. J Physiol. 2016. PMID: 26574187 Free PMC article.
References
-
- Thornton D. J., Rousseau K., McGuckin M. A. (2008) Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 70, 459–486 - PubMed
-
- Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. (2002) Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 - PubMed
-
- Boucher R. C. (2004) New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23, 146–158 - PubMed
-
- Rogers D. F. (2004) Airway mucus hypersecretion in asthma: an undervalued pathology? Curr. Opin. Pharmacol. 4, 241–250 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

