Bacterial scaffold directs pole-specific centromere segregation
- PMID: 24778223
- PMCID: PMC4024888
- DOI: 10.1073/pnas.1405188111
Bacterial scaffold directs pole-specific centromere segregation
Abstract
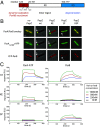
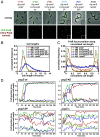
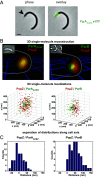
Bacteria use partitioning systems based on the ParA ATPase to actively mobilize and spatially organize molecular cargoes throughout the cytoplasm. The bacterium Caulobacter crescentus uses a ParA-based partitioning system to segregate newly replicated chromosomal centromeres to opposite cell poles. Here we demonstrate that the Caulobacter PopZ scaffold creates an organizing center at the cell pole that actively regulates polar centromere transport by the ParA partition system. As segregation proceeds, the ParB-bound centromere complex is moved by progressively disassembling ParA from a nucleoid-bound structure. Using superresolution microscopy, we show that released ParA is recruited directly to binding sites within a 3D ultrastructure composed of PopZ at the cell pole, whereas the ParB-centromere complex remains at the periphery of the PopZ structure. PopZ recruitment of ParA stimulates ParA to assemble on the nucleoid near the PopZ-proximal cell pole. We identify mutations in PopZ that allow scaffold assembly but specifically abrogate interactions with ParA and demonstrate that PopZ/ParA interactions are required for proper chromosome segregation in vivo. We propose that during segregation PopZ sequesters free ParA and induces target-proximal regeneration of ParA DNA binding activity to enforce processive and pole-directed centromere segregation, preventing segregation reversals. PopZ therefore functions as a polar hub complex at the cell pole to directly regulate the directionality and destination of transfer of the mitotic segregation machine.
Keywords: parAB; prokaryotic; replication; soj; spo0J.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Polar Organizing Protein PopZ Is Required for Chromosome Segregation in Agrobacterium tumefaciens.J Bacteriol. 2017 Aug 8;199(17):e00111-17. doi: 10.1128/JB.00111-17. Print 2017 Sep 1. J Bacteriol. 2017. PMID: 28630129 Free PMC article.
-
To let go or not to let go: how ParA can impact the release of the chromosomal anchoring in Caulobacter crescentus.Nucleic Acids Res. 2023 Dec 11;51(22):12275-12287. doi: 10.1093/nar/gkad982. Nucleic Acids Res. 2023. PMID: 37933842 Free PMC article.
-
A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole.Cell. 2008 Sep 19;134(6):945-55. doi: 10.1016/j.cell.2008.07.015. Cell. 2008. PMID: 18805088 Free PMC article.
-
End-in-Sight: Cell Polarization by the Polygamic Organizer PopZ.Trends Microbiol. 2018 Apr;26(4):363-375. doi: 10.1016/j.tim.2017.11.007. Epub 2017 Nov 29. Trends Microbiol. 2018. PMID: 29198650 Review.
-
Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle.Plasmid. 2012 Jan;67(1):1-14. doi: 10.1016/j.plasmid.2011.08.003. Epub 2011 Sep 6. Plasmid. 2012. PMID: 21924286 Review.
Cited by
-
Rapid pairing and resegregation of distant homologous loci enables double-strand break repair in bacteria.J Cell Biol. 2015 Aug 3;210(3):385-400. doi: 10.1083/jcb.201505019. J Cell Biol. 2015. PMID: 26240183 Free PMC article.
-
The Polar Organizing Protein PopZ Is Fundamental for Proper Cell Division and Segregation of Cellular Content in Magnetospirillum gryphiswaldense.mBio. 2019 Mar 12;10(2):e02716-18. doi: 10.1128/mBio.02716-18. mBio. 2019. PMID: 30862753 Free PMC article.
-
PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11666-71. doi: 10.1073/pnas.1515544112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324921 Free PMC article.
-
Mechanism for Anaphase B: Evaluation of "Slide-and-Cluster" versus "Slide-and-Flux-or-Elongate" Models.Biophys J. 2015 Apr 21;108(8):2007-18. doi: 10.1016/j.bpj.2015.03.018. Biophys J. 2015. PMID: 25902440 Free PMC article.
-
Crosstalk Regulation Between Bacterial Chromosome Replication and Chromosome Partitioning.Front Microbiol. 2019 Feb 26;10:279. doi: 10.3389/fmicb.2019.00279. eCollection 2019. Front Microbiol. 2019. PMID: 30863373 Free PMC article. Review.
References
-
- Nevo-Dinur K, Govindarajan S, Amster-Choder O. Subcellular localization of RNA and proteins in prokaryotes. Trends Genet. 2012;28(7):314–322. - PubMed
-
- Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol. 2006;61(5):1352–1361. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

