A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria
- PMID: 24778229
- PMCID: PMC4024899
- DOI: 10.1073/pnas.1402358111
A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria
Abstract
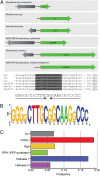
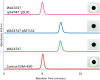
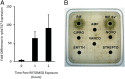
Many environmental bacteria are multidrug-resistant and represent a reservoir of ancient antibiotic resistance determinants, which have been linked to genes found in pathogens. Exploring the environmental antibiotic resistome, therefore, reveals the diversity and evolution of antibiotic resistance and also provides insight into the vulnerability of clinically used antibiotics. In this study, we describe the identification of a highly conserved regulatory motif, the rifampin (RIF) -associated element (RAE), which is found upstream of genes encoding RIF-inactivating enzymes from a diverse collection of actinomycetes. Using gene expression assays, we confirmed that the RAE is involved in RIF-responsive regulation. By using the RAE as a probe for new RIF-associated genes in several actinomycete genomes, we identified a heretofore unknown RIF resistance gene, RIF phosphotransferase (rph). The RPH enzyme is a RIF-inactivating phosphotransferase and represents a new protein family in antibiotic resistance. RPH orthologs are widespread and found in RIF-sensitive bacteria, including Bacillus cereus and the pathogen Listeria monocytogenes. Heterologous expression and in vitro enzyme assays with purified RPHs from diverse bacterial genera show that these enzymes are capable of conferring high-level resistance to a variety of clinically used rifamycin antibiotics. This work identifies a new antibiotic resistance protein family and reinforces the fact that the study of resistance in environmental organisms can serve to identify resistance elements with relevance to pathogens.
Keywords: drug inactivation; gene regulation; phosphorylation; silent resistome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Rifampin phosphotransferase is an unusual antibiotic resistance kinase.Nat Commun. 2016 Apr 22;7:11343. doi: 10.1038/ncomms11343. Nat Commun. 2016. PMID: 27103605 Free PMC article.
-
The Enzymes of the Rifamycin Antibiotic Resistome.Acc Chem Res. 2021 May 4;54(9):2065-2075. doi: 10.1021/acs.accounts.1c00048. Epub 2021 Apr 20. Acc Chem Res. 2021. PMID: 33877820 Review.
-
Structural basis of rifampin inactivation by rifampin phosphotransferase.Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):3803-8. doi: 10.1073/pnas.1523614113. Epub 2016 Mar 21. Proc Natl Acad Sci U S A. 2016. PMID: 27001859 Free PMC article.
-
Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes.Antimicrob Agents Chemother. 2012 Oct;56(10):5061-9. doi: 10.1128/AAC.01166-12. Epub 2012 Jul 16. Antimicrob Agents Chemother. 2012. PMID: 22802246 Free PMC article.
-
The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics.J Appl Microbiol. 2016 Feb;120(2):251-65. doi: 10.1111/jam.12989. Epub 2015 Dec 28. J Appl Microbiol. 2016. PMID: 26509460 Review.
Cited by
-
Rifampin phosphotransferase is an unusual antibiotic resistance kinase.Nat Commun. 2016 Apr 22;7:11343. doi: 10.1038/ncomms11343. Nat Commun. 2016. PMID: 27103605 Free PMC article.
-
Bacterial Enzymes and Antibiotic Resistance.Acta Naturae. 2018 Oct-Dec;10(4):33-48. Acta Naturae. 2018. PMID: 30713760 Free PMC article.
-
Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3')-VI.mBio. 2014 Oct 21;5(5):e01972-14. doi: 10.1128/mBio.01972-14. mBio. 2014. PMID: 25336457 Free PMC article.
-
Molecular mechanisms of antibiotic resistance revisited.Nat Rev Microbiol. 2023 May;21(5):280-295. doi: 10.1038/s41579-022-00820-y. Epub 2022 Nov 21. Nat Rev Microbiol. 2023. PMID: 36411397 Review.
-
Host and gut bacteria share metabolic pathways for anti-cancer drug metabolism.Nat Microbiol. 2022 Oct;7(10):1605-1620. doi: 10.1038/s41564-022-01226-5. Epub 2022 Sep 22. Nat Microbiol. 2022. PMID: 36138165 Free PMC article.
References
-
- Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472(7341):32. - PubMed
-
- Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot (Tokyo) 2012;65(8):441. - PubMed
-
- D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. - PubMed
-
- D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311(5759):374–377. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

