Syntrophic exchange in synthetic microbial communities
- PMID: 24778240
- PMCID: PMC4034247
- DOI: 10.1073/pnas.1405641111
Syntrophic exchange in synthetic microbial communities
Abstract
Metabolic crossfeeding is an important process that can broadly shape microbial communities. However, little is known about specific crossfeeding principles that drive the formation and maintenance of individuals within a mixed population. Here, we devised a series of synthetic syntrophic communities to probe the complex interactions underlying metabolic exchange of amino acids. We experimentally analyzed multimember, multidimensional communities of Escherichia coli of increasing sophistication to assess the outcomes of synergistic crossfeeding. We find that biosynthetically costly amino acids including methionine, lysine, isoleucine, arginine, and aromatics, tend to promote stronger cooperative interactions than amino acids that are cheaper to produce. Furthermore, cells that share common intermediates along branching pathways yielded more synergistic growth, but exhibited many instances of both positive and negative epistasis when these interactions scaled to higher dimensions. In more complex communities, we find certain members exhibiting keystone species-like behavior that drastically impact the community dynamics. Based on comparative genomic analysis of >6,000 sequenced bacteria from diverse environments, we present evidence suggesting that amino acid biosynthesis has been broadly optimized to reduce individual metabolic burden in favor of enhanced crossfeeding to support synergistic growth across the biosphere. These results improve our basic understanding of microbial syntrophy while also highlighting the utility and limitations of current modeling approaches to describe the dynamic complexities underlying microbial ecosystems. This work sets the foundation for future endeavors to resolve key questions in microbial ecology and evolution, and presents a platform to develop better and more robust engineered synthetic communities for industrial biotechnology.
Keywords: amino acid exchange; population modeling; synthetic ecosystem.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


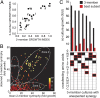

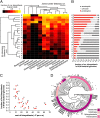
References
-
- Brenner K, You L, Arnold FH. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008;26(9):483–489. - PubMed
-
- Shong J, Jimenez Diaz MR, Collins CH. Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol. 2012;23(5):798–802. - PubMed
-
- Bayer TS, et al. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131(18):6508–6515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

