Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia
- PMID: 24778245
- PMCID: PMC4024891
- DOI: 10.1073/pnas.1313093111
Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia
Abstract
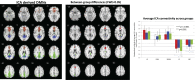
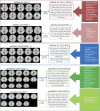
The brain's default mode network (DMN) is highly heritable and is compromised in a variety of psychiatric disorders. However, genetic control over the DMN in schizophrenia (SZ) and psychotic bipolar disorder (PBP) is largely unknown. Study subjects (n = 1,305) underwent a resting-state functional MRI scan and were analyzed by a two-stage approach. The initial analysis used independent component analysis (ICA) in 324 healthy controls, 296 SZ probands, 300 PBP probands, 179 unaffected first-degree relatives of SZ probands (SZREL), and 206 unaffected first-degree relatives of PBP probands to identify DMNs and to test their biomarker and/or endophenotype status. A subset of controls and probands (n = 549) then was subjected to a parallel ICA (para-ICA) to identify imaging-genetic relationships. ICA identified three DMNs. Hypo-connectivity was observed in both patient groups in all DMNs. Similar patterns observed in SZREL were restricted to only one network. DMN connectivity also correlated with several symptom measures. Para-ICA identified five sub-DMNs that were significantly associated with five different genetic networks. Several top-ranking SNPs across these networks belonged to previously identified, well-known psychosis/mood disorder genes. Global enrichment analyses revealed processes including NMDA-related long-term potentiation, PKA, immune response signaling, axon guidance, and synaptogenesis that significantly influenced DMN modulation in psychoses. In summary, we observed both unique and shared impairments in functional connectivity across the SZ and PBP cohorts; these impairments were selectively familial only for SZREL. Genes regulating specific neurodevelopment/transmission processes primarily mediated DMN disconnectivity. The study thus identifies biological pathways related to a widely researched quantitative trait that might suggest novel, targeted drug treatments for these diseases.
Keywords: BSNIP; architecture; genetics; molecular.
Conflict of interest statement
Conflict of interest statement: J.A.S. receives support from Janssen, Takeda, Bristol-Myers Squibb, Roche, and Lilly. G.P. has been a consultant for Bristol-Myers Squibb. G.R. is the president and A.W. is a former employee of Genomas Inc. M.S.K. has received support from Sunovion.
Figures


References
-
- Ellison-Wright I, Bullmore E. Anatomy of bipolar disorder and schizophrenia: A meta-analysis. Schizophr Res. 2010;117(1):1–12. - PubMed
Publication types
MeSH terms
Grants and funding
- MH077862/MH/NIMH NIH HHS/United States
- MH077852/MH/NIMH NIH HHS/United States
- R01 MH077862/MH/NIMH NIH HHS/United States
- MH078113/MH/NIMH NIH HHS/United States
- MH077945/MH/NIMH NIH HHS/United States
- R44 MH075481/MH/NIMH NIH HHS/United States
- R01 MH077851/MH/NIMH NIH HHS/United States
- R01 MH078113/MH/NIMH NIH HHS/United States
- R01 MH077852/MH/NIMH NIH HHS/United States
- MH077851/MH/NIMH NIH HHS/United States
- 2R44 MH075481/MH/NIMH NIH HHS/United States
- R01 MH096913/MH/NIMH NIH HHS/United States
- R01 MH077945/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

