A novel Netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching
- PMID: 24778312
- PMCID: PMC4003241
- DOI: 10.1083/jcb.201311003
A novel Netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching
Abstract
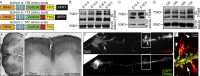
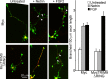
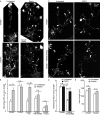
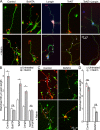
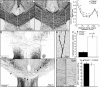
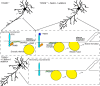
Developmental axon branching dramatically increases synaptic capacity and neuronal surface area. Netrin-1 promotes branching and synaptogenesis, but the mechanism by which Netrin-1 stimulates plasma membrane expansion is unknown. We demonstrate that SNARE-mediated exocytosis is a prerequisite for axon branching and identify the E3 ubiquitin ligase TRIM9 as a critical catalytic link between Netrin-1 and exocytic SNARE machinery in murine cortical neurons. TRIM9 ligase activity promotes SNARE-mediated vesicle fusion and axon branching in a Netrin-dependent manner. We identified a direct interaction between TRIM9 and the Netrin-1 receptor DCC as well as a Netrin-1-sensitive interaction between TRIM9 and the SNARE component SNAP25. The interaction with SNAP25 negatively regulates SNARE-mediated exocytosis and axon branching in the absence of Netrin-1. Deletion of TRIM9 elevated exocytosis in vitro and increased axon branching in vitro and in vivo. Our data provide a novel model for the spatial regulation of axon branching by Netrin-1, in which localized plasma membrane expansion occurs via TRIM9-dependent regulation of SNARE-mediated vesicle fusion.
Figures



References
-
- Bouchard J.-F., Moore S.W., Tritsch N.X., Roux P.P., Shekarabi M., Barker P.A., Kennedy T.E. 2004. Protein kinase A activation promotes plasma membrane insertion of DCC from an intracellular pool: A novel mechanism regulating commissural axon extension. J. Neurosci. 24:3040–3050 10.1523/JNEUROSCI.4934-03.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

