Delayed transition to new cell fates during cellular reprogramming
- PMID: 24780626
- PMCID: PMC4064802
- DOI: 10.1016/j.ydbio.2014.04.015
Delayed transition to new cell fates during cellular reprogramming
Abstract
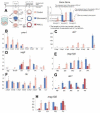
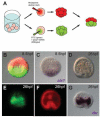
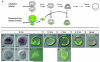
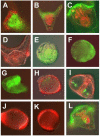
In many embryos specification toward one cell fate can be diverted to a different cell fate through a reprogramming process. Understanding how that process works will reveal insights into the developmental regulatory logic that emerged from evolution. In the sea urchin embryo, cells at gastrulation were found to reprogram and replace missing cell types after surgical dissections of the embryo. Non-skeletogenic mesoderm (NSM) cells reprogrammed to replace missing skeletogenic mesoderm cells and animal caps reprogrammed to replace all endomesoderm. In both cases evidence of reprogramming onset was first observed at the early gastrula stage, even if the cells to be replaced were removed earlier in development. Once started however, the reprogramming occurred with compressed gene expression dynamics. The NSM did not require early contact with the skeletogenic cells to reprogram, but the animal cap cells gained the ability to reprogram early in gastrulation only after extended contact with the vegetal halves prior to that time. If the entire vegetal half was removed at early gastrula, the animal caps reprogrammed and replaced the vegetal half endomesoderm. If the animal caps carried morpholinos to either hox11/13b or foxA (endomesoderm specification genes), the isolated animal caps failed to reprogram. Together these data reveal that the emergence of a reprogramming capability occurs at early gastrulation in the sea urchin embryo and requires activation of early specification components of the target tissues.
Keywords: Cell fate; Cell fate specification; Differentiation; Gene regulatory network; Regulative development; Reprogramming; Sea urchin embryo.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Amemiya S. Complete regulation of development throughout metamorphosis of sea urchin embryos devoid of macromeres. Dev Growth Differ. 1996;38:465–476. - PubMed
-
- Davidson EH. Academic; Burlington, MA; San Diego: 2006. The regulatory genome: gene regulatory networks in development and evolution.
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

