ABORTED MICROSPORES Acts as a Master Regulator of Pollen Wall Formation in Arabidopsis
- PMID: 24781116
- PMCID: PMC4036570
- DOI: 10.1105/tpc.114.122986
ABORTED MICROSPORES Acts as a Master Regulator of Pollen Wall Formation in Arabidopsis
Abstract
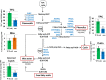
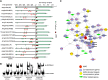
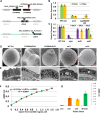
Mature pollen is covered by durable cell walls, principally composed of sporopollenin, an evolutionary conserved, highly resilient, but not fully characterized, biopolymer of aliphatic and aromatic components. Here, we report that ABORTED MICROSPORES (AMS) acts as a master regulator coordinating pollen wall development and sporopollenin biosynthesis in Arabidopsis thaliana. Genome-wide coexpression analysis revealed 98 candidate genes with specific expression in the anther and 70 that showed reduced expression in ams. Among these 70 members, we showed that AMS can directly regulate 23 genes implicated in callose dissociation, fatty acids elongation, formation of phenolic compounds, and lipidic transport putatively involved in sporopollenin precursor synthesis. Consistently, ams mutants showed defective microspore release, a lack of sporopollenin deposition, and a dramatic reduction in total phenolic compounds and cutin monomers. The functional importance of the AMS pathway was further demonstrated by the observation of impaired pollen wall architecture in plant lines with reduced expression of several AMS targets: the abundant pollen coat protein extracellular lipases (EXL5 and EXL6), and CYP98A8 and CYP98A9, which are enzymes required for the production of phenolic precursors. These findings demonstrate the central role of AMS in coordinating sporopollenin biosynthesis and the secretion of materials for pollen wall patterning.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Ahlers F., Thom I., Lambert J., Kuckuk R., Rolf W. (1999). 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 50: 1095–1098
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Alves Ferreira M., de Almeida Engler J., Costa Miguens F., Van Montagu M., Engler G., de Oliveira D.E. (1997). Oleosin gene expression in Arabidopsis thaliana tapetum coincides with accumulation of lipids in plastids and cytoplasmic bodies. Plant Physiol. Biochem. 35: 729–739
-
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460 - PubMed
-
- Blackmore S., Wortley A.H., Skvarla J.J., Rowley J.R. (2007). Pollen wall development in flowering plants. New Phytol. 174: 483–498 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

