Geographic population structure analysis of worldwide human populations infers their biogeographical origins
- PMID: 24781250
- PMCID: PMC4007635
- DOI: 10.1038/ncomms4513
Geographic population structure analysis of worldwide human populations infers their biogeographical origins
Erratum in
-
Corrigendum: Geographic population structure analysis of worldwide human populations infers their biogeographical origins.Nat Commun. 2016 Oct 31;7:13468. doi: 10.1038/ncomms13468. Nat Commun. 2016. PMID: 27796289 Free PMC article. No abstract available.
Abstract
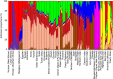
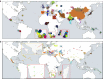
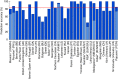
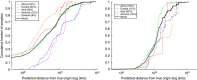
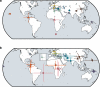
The search for a method that utilizes biological information to predict humans' place of origin has occupied scientists for millennia. Over the past four decades, scientists have employed genetic data in an effort to achieve this goal but with limited success. While biogeographical algorithms using next-generation sequencing data have achieved an accuracy of 700 km in Europe, they were inaccurate elsewhere. Here we describe the Geographic Population Structure (GPS) algorithm and demonstrate its accuracy with three data sets using 40,000-130,000 SNPs. GPS placed 83% of worldwide individuals in their country of origin. Applied to over 200 Sardinians villagers, GPS placed a quarter of them in their villages and most of the rest within 50 km of their villages. GPS's accuracy and power to infer the biogeography of worldwide individuals down to their country or, in some cases, village, of origin, underscores the promise of admixture-based methods for biogeography and has ramifications for genetic ancestry testing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Tishkoff S. A. & Kidd K. K. Implications of biogeography of human populations for ‘race’ and medicine. Nat. Genet. 36, S21–S27 (2004). - PubMed
-
- Harcourt A. H. Human Biogeography University of California Press (2012).
-
- Darwin C. The Descent of Man and Selection in Relation to Sex John Murray London (1871).
-
- Rowe J. H. The Renaissance Foundations of Anthropology. American Anthropologist 67, 1–20 (1965).
-
- Cavalli-Sforza L. L. L., Menozzi P. & Piazza A. The History and Geography of Human Genes Princeton university press (1994).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

