A multi-fingerprint browser for the ZINC database
- PMID: 24782520
- PMCID: PMC4086083
- DOI: 10.1093/nar/gku379
A multi-fingerprint browser for the ZINC database
Abstract
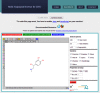
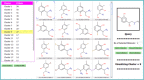
To confirm the activity of an initial small molecule 'hit compound' from an activity screening, one needs to probe the structure-activity relationships by testing close analogs. The multi-fingerprint browser presented here (http://dcb-reymond23.unibe.ch:8080/MCSS/) enables one to rapidly identify such close analogs among commercially available compounds in the ZINC database (>13 million molecules). The browser retrieves nearest neighbors of any query molecule in multi-dimensional chemical spaces defined by four different fingerprints, each of which represents relevant structural and pharmacophoric features in a different way: sFP (substructure fingerprint), ECFP4 (extended connectivity fingerprint), MQNs (molecular quantum numbers) and SMIfp (SMILES fingerprint). Distances are calculated using the city-block distance, a similarity measure that performs as well as Tanimoto similarity but is much faster to compute. The list of up to 1000 nearest neighbors of any query molecule is retrieved by the browser and can be then clustered using the K-means clustering algorithm to produce a focused list of analogs with likely similar bioactivity to be considered for experimental evaluation.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bleicher K.H., Bohm H.-J., Muller K., Alanine A.I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug. Discov. 2003;2:369–378. - PubMed
-
- Ripphausen P., Nisius B., Peltason L., Bajorath J. Quo vadis, virtual screening? A comprehensive survey of prospective applications. J. Med. Chem. 2010;53:8461–8467. - PubMed
-
- Geppert H., Vogt M., Bajorath J. Current trends in ligand-based virtual screening: molecular representations, data mining methods, new application areas, and performance evaluation. J. Chem. Inf. Model. 2010;50:205–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

