Machine learning patterns for neuroimaging-genetic studies in the cloud
- PMID: 24782753
- PMCID: PMC3986524
- DOI: 10.3389/fninf.2014.00031
Machine learning patterns for neuroimaging-genetic studies in the cloud
Abstract
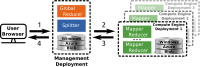
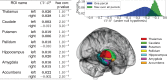
Brain imaging is a natural intermediate phenotype to understand the link between genetic information and behavior or brain pathologies risk factors. Massive efforts have been made in the last few years to acquire high-dimensional neuroimaging and genetic data on large cohorts of subjects. The statistical analysis of such data is carried out with increasingly sophisticated techniques and represents a great computational challenge. Fortunately, increasing computational power in distributed architectures can be harnessed, if new neuroinformatics infrastructures are designed and training to use these new tools is provided. Combining a MapReduce framework (TomusBLOB) with machine learning algorithms (Scikit-learn library), we design a scalable analysis tool that can deal with non-parametric statistics on high-dimensional data. End-users describe the statistical procedure to perform and can then test the model on their own computers before running the very same code in the cloud at a larger scale. We illustrate the potential of our approach on real data with an experiment showing how the functional signal in subcortical brain regions can be significantly fit with genome-wide genotypes. This experiment demonstrates the scalability and the reliability of our framework in the cloud with a 2 weeks deployment on hundreds of virtual machines.
Keywords: cloud computing; fMRI; heritability; machine learning; neuroimaging-genetic.
Figures

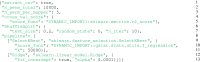
References
-
- Anderson M. J., Robinson J. (2001). Permutation tests for linear models. Aust. N. Z. J. Stat. 43, 75–88 10.1111/1467-842X.00156 - DOI
-
- Chu C.-T., Kim S. K., Lin Y.-A., Yu Y., Bradski G. R., Ng A. Y., et al. (2006). Map-reduce for machine learning on multicore, in NIPS (Vancouver, BC: ), 281–288
-
- Da Mota B., Frouin V., Duchesnay E., Laguitton S., Varoquaux G., Poline J.-B., et al. (2012). “A fast computational framework for genome-wide association studies with neuroimaging data,” in 20th International Conference on Computational Statistics (Limassol: ).
LinkOut - more resources
Full Text Sources
Other Literature Sources

