Glutamine synthetase 2 is not essential for biosynthesis of compatible solutes in Halobacillus halophilus
- PMID: 24782854
- PMCID: PMC3995056
- DOI: 10.3389/fmicb.2014.00168
Glutamine synthetase 2 is not essential for biosynthesis of compatible solutes in Halobacillus halophilus
Abstract
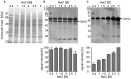
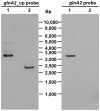
Halobacillus halophilus, a moderately halophilic bacterium isolated from salt marshes, produces various compatible solutes to cope with osmotic stress. Glutamate and glutamine are dominant compatible solutes at mild salinities. Glutamine synthetase activity in cell suspensions of Halobacillus halophilus wild type was shown to be salt dependent and chloride modulated. A possible candidate to catalyze glutamine synthesis is glutamine synthetase A2, whose transcription is stimulated by chloride. To address the role of GlnA2 in the biosynthesis of the osmolytes glutamate and glutamine, a deletion mutant (ΔglnA2) was generated and characterized in detail. We compared the pool of compatible solutes and performed transcriptional analyses of the principal genes controlling the solute production in the wild type strain and the deletion mutant. These measurements did not confirm the hypothesized role of GlnA2 in the osmolyte production. Most likely the presence of another, yet to be identified enzyme has the main contribution in the measured activity in crude extracts and probably determines the total chloride-modulated profile. The role of GlnA2 remains to be elucidated.
Keywords: Halobacillus halophilus; compatible solutes; glutamine synthetase; halophile; osmoregulation.
Figures

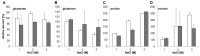



References
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (1992). Current Protocols in Molecular Biology. New York: Green Publishing and Wiley-Interscience; 10.1002/0471142727 - DOI
-
- Cánovas D., Vargas C., Calderon M. I., Ventosa A., Nieto J. J. (1998). Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. Syst. Appl. Microbiol. 21 487–497 10.1016/S0723-2020(98)80060-X - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

