Global Regulation of Embryonic Patterning in Arabidopsis by MicroRNAs
- PMID: 24784759
- PMCID: PMC4044841
- DOI: 10.1104/pp.114.240846
Global Regulation of Embryonic Patterning in Arabidopsis by MicroRNAs
Abstract
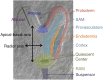
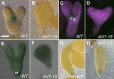
The development of the embryo in Arabidopsis (Arabidopsis thaliana) involves a carefully controlled set of cell divisions and cell fate decisions that lead to a mature embryo containing shoot and root meristems and all basic tissue types. Over the last 20 years, a number of transcriptional regulators of embryonic patterning have been described, but little is known about the role of posttranscriptional regulators such as microRNAs (miRNAs). Previous work has centered on the study of null or very weak alleles of miRNA biosynthetic genes, but these mutants either arrest early in embryogenesis or have wild-type-looking embryos. Here, we significantly extend those analyses by characterizing embryos mutant for a strong hypomorphic allele of DICER-LIKE1 (dcl1-15). Our data demonstrate that miRNAs are required for the patterning of most regions of the embryo, with the exception of the protoderm. In mutant embryos with the most severe morphological defects, the majority of tissue identities are lost. Different levels of miRNAs appear to be required to specify cell fates in various regions of the embryo. The suspensor needs the lowest levels, followed by the root apical meristem and hypocotyl, cotyledons, and shoot apical meristem. Furthermore, we show that erecta acts as a suppressor of dcl1-15, a novel role for this signaling pathway in embryos. Our results also indicate that the regulation of the messenger RNA levels of miRNA targets involves not just the action of miRNAs but has a significant transcriptional component as well.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures






Similar articles
-
POLTERGEIST and POLTERGEIST-LIKE1 are essential for the maintenance of post-embryonic shoot and root apical meristems as revealed by a partial loss-of-function mutant allele of pll1 in Arabidopsis.Genes Genomics. 2020 Jan;42(1):107-116. doi: 10.1007/s13258-019-00894-8. Epub 2019 Dec 3. Genes Genomics. 2020. PMID: 31797316
-
Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote.Dev Biol. 2017 Nov 15;431(2):145-151. doi: 10.1016/j.ydbio.2017.09.009. Epub 2017 Sep 12. Dev Biol. 2017. PMID: 28912016
-
MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis.Genes Dev. 2010 Dec 1;24(23):2678-92. doi: 10.1101/gad.1986710. Genes Dev. 2010. PMID: 21123653 Free PMC article.
-
Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo.J Exp Bot. 2011 Mar;62(5):1687-97. doi: 10.1093/jxb/erq398. Epub 2010 Dec 20. J Exp Bot. 2011. PMID: 21172809 Review.
-
Expression of microRNAs and its regulation in plants.Semin Cell Dev Biol. 2010 Oct;21(8):790-7. doi: 10.1016/j.semcdb.2010.03.012. Epub 2010 Apr 18. Semin Cell Dev Biol. 2010. PMID: 20403450 Free PMC article. Review.
Cited by
-
Protocols for Obtaining Zygotic and Somatic Embryos for Studying the Regulation of Early Embryo Development in the Model Legume Medicago truncatula.J Vis Exp. 2015 Jun 9;(100):e52635. doi: 10.3791/52635. J Vis Exp. 2015. PMID: 26131626 Free PMC article.
-
MicroRNA Dynamics and Functions During Arabidopsis Embryogenesis.Plant Cell. 2019 Dec;31(12):2929-2946. doi: 10.1105/tpc.19.00395. Epub 2019 Sep 27. Plant Cell. 2019. PMID: 31562217 Free PMC article.
-
Identification of MicroRNAs and their Targets Associated with Embryo Abortion during Chrysanthemum Cross Breeding via High-Throughput Sequencing.PLoS One. 2015 Apr 24;10(4):e0124371. doi: 10.1371/journal.pone.0124371. eCollection 2015. PLoS One. 2015. PMID: 25909659 Free PMC article.
-
Gene expression atlas of embryo development in Arabidopsis.Plant Reprod. 2019 Mar;32(1):93-104. doi: 10.1007/s00497-019-00364-x. Epub 2019 Feb 14. Plant Reprod. 2019. PMID: 30762127
-
The explant developmental stage profoundly impacts small RNA-mediated regulation at the dedifferentiation step of maize somatic embryogenesis.Sci Rep. 2019 Oct 10;9(1):14511. doi: 10.1038/s41598-019-50962-y. Sci Rep. 2019. PMID: 31601893 Free PMC article.
References
-
- Abe M, Katsumata H, Komeda Y, Takahashi T. (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 - PubMed
-
- Aida M, Ishida T, Tasaka M. (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 - PubMed
-
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

