Copy number variation is a fundamental aspect of the placental genome
- PMID: 24785991
- PMCID: PMC4006706
- DOI: 10.1371/journal.pgen.1004290
Copy number variation is a fundamental aspect of the placental genome
Abstract
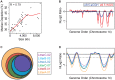
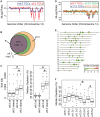
Discovery of lineage-specific somatic copy number variation (CNV) in mammals has led to debate over whether CNVs are mutations that propagate disease or whether they are a normal, and even essential, aspect of cell biology. We show that 1,000 N polyploid trophoblast giant cells (TGCs) of the mouse placenta contain 47 regions, totaling 138 Megabases, where genomic copies are underrepresented (UR). UR domains originate from a subset of late-replicating heterochromatic regions containing gene deserts and genes involved in cell adhesion and neurogenesis. While lineage-specific CNVs have been identified in mammalian cells, classically in the immune system where V(D)J recombination occurs, we demonstrate that CNVs form during gestation in the placenta by an underreplication mechanism, not by recombination nor deletion. Our results reveal that large scale CNVs are a normal feature of the mammalian placental genome, which are regulated systematically during embryogenesis and are propagated by a mechanism of underreplication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

