Key tumor suppressor genes inactivated by "greater promoter" methylation and somatic mutations in head and neck cancer
- PMID: 24786473
- PMCID: PMC4143405
- DOI: 10.4161/epi.29025
Key tumor suppressor genes inactivated by "greater promoter" methylation and somatic mutations in head and neck cancer
Abstract
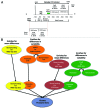
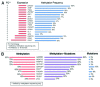
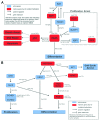
Tumor suppressor genes (TSGs) are commonly inactivated by somatic mutation and/or promoter methylation; yet, recent high-throughput genomic studies have not identified key TSGs inactivated by both mechanisms. We pursued an integrated molecular analysis based on methylation binding domain sequencing (MBD-seq), 450K Methylation arrays, whole exome sequencing, and whole genome gene expression arrays in primary head and neck squamous cell carcinoma (HNSCC) tumors and matched uvulopalatopharyngoplasty tissue samples (UPPPs). We uncovered 186 downregulated genes harboring cancer specific promoter methylation including PAX1 and PAX5 and we identified 10 key tumor suppressor genes (GABRB3, HOXC12, PARP15, SLCO4C1, CDKN2A, PAX1, PIK3AP1, HOXC6, PLCB1, and ZIC4) inactivated by both promoter methylation and/or somatic mutation. Among the novel tumor suppressor genes discovered with dual mechanisms of inactivation, we found a high frequency of genomic and epigenomic alterations in the PAX gene family of transcription factors, which selectively impact canonical NOTCH and TP53 pathways to determine cell fate, cell survival, and genome maintenance. Our results highlight the importance of assessing TSGs at the genomic and epigenomic level to identify key pathways in HNSCC, deregulated by simultaneous promoter methylation and somatic mutations.
Keywords: DNA methylation; Head and Neck Squamous Cell Carcinoma; Tumor Suppressor Genes; integration analysis; somatic mutations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
