Stability of bivariate GWAS biomarker detection
- PMID: 24787002
- PMCID: PMC4005767
- DOI: 10.1371/journal.pone.0093319
Stability of bivariate GWAS biomarker detection
Abstract
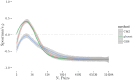
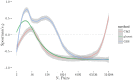
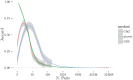
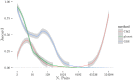
Given the difficulty and effort required to confirm candidate causal SNPs detected in genome-wide association studies (GWAS), there is no practical way to definitively filter false positives. Recent advances in algorithmics and statistics have enabled repeated exhaustive search for bivariate features in a practical amount of time using standard computational resources, allowing us to use cross-validation to evaluate the stability. We performed 10 trials of 2-fold cross-validation of exhaustive bivariate analysis on seven Wellcome-Trust Case-Control Consortium GWAS datasets, comparing the traditional [Formula: see text] test for association, the high-performance GBOOST method and the recently proposed GSS statistic (Available at http://bioinformatics.research.nicta.com.au/software/gwis/). We use Spearman's correlation to measure the similarity between the folds of cross validation. To compare incomplete lists of ranks we propose an extension to Spearman's correlation. The extension allows us to consider a natural threshold for feature selection where the correlation is zero. This is the first reported cross-validation study of exhaustive bivariate GWAS feature selection. We found that stability between ranked lists from different cross-validation folds was higher for GSS in the majority of diseases. A thorough analysis of the correlation between SNP-frequency and univariate [Formula: see text] score demonstrated that the [Formula: see text] test for association is highly confounded by main effects: SNPs with high univariate significance replicably dominate the ranked results. We show that removal of the univariately significant SNPs improves [Formula: see text] replicability but risks filtering pairs involving SNPs with univariate effects. We empirically confirm that the stability of GSS and GBOOST were not affected by removal of univariately significant SNPs. These results suggest that the GSS and GBOOST tests are successfully targeting bivariate association with phenotype and that GSS is able to reliably detect a larger set of SNP-pairs than GBOOST in the majority of the data we analysed. However, the [Formula: see text] test for association was confounded by main effects.
Conflict of interest statement
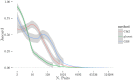
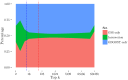
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

