Processing of heparanase is mediated by syndecan-1 cytoplasmic domain and involves syntenin and α-actinin
- PMID: 24788042
- PMCID: PMC4209210
- DOI: 10.1007/s00018-014-1629-9
Processing of heparanase is mediated by syndecan-1 cytoplasmic domain and involves syntenin and α-actinin
Abstract
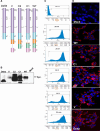
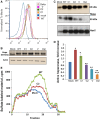
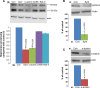
Heparanase activity plays a decisive role in cell dissemination associated with cancer metastasis. Cellular uptake of heparanase is considered a pre-requisite for the delivery of latent 65-kDa heparanase to lysosomes and its subsequent proteolytic processing and activation into 8- and 50-kDa protein subunits by cathepsin L. Heparan sulfate proteoglycans, and particularly syndecan, are instrumental for heparanase uptake and activation, through a process that has been shown to occur independent of rafts. Nevertheless, the molecular mechanism underlying syndecan-mediated internalization outside of rafts is unclear. Here, we examined the role of syndecan-1 cytoplasmic domain in heparanase processing, utilizing deletion constructs lacking the entire cytoplasmic domain (Delta), the conserved (C1 or C2), or variable (V) regions. Heparanase processing was markedly increased following syndecan-1 over-expression; in contrast, heparanase was retained at the cell membrane and its processing was impaired in cells over-expressing syndecan-1 deleted for the entire cytoplasmic tail. We have next revealed that conserved domain 2 (C2) and variable (V) regions of syndecan-1 cytoplasmic tail mediate heparanase processing. Furthermore, we found that syntenin, known to interact with syndecan C2 domain, and α actinin are essential for heparanase processing.
Figures



References
-
- Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

