Time course transcriptome changes in Shewanella algae in response to salt stress
- PMID: 24789066
- PMCID: PMC4006864
- DOI: 10.1371/journal.pone.0096001
Time course transcriptome changes in Shewanella algae in response to salt stress
Abstract
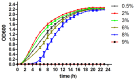
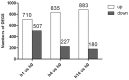
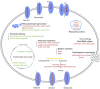
Shewanella algae, which produces tetrodotoxin and exists in various seafoods, can cause human diseases, such as spondylodiscitis and bloody diarrhea. In the present study, we focused on the temporal, dynamic process in salt-stressed S. algae by monitoring the gene transcript levels at different time points after high salt exposure. Transcript changes in amino acid metabolism, carbohydrate metabolism, energy metabolism, membrane transport, regulatory functions, and cellular signaling were found to be important for the high salt response in S. algae. The most common strategies used by bacteria to survive and grow in high salt environments, such as Na+ efflux, K+ uptake, glutamate transport and biosynthesis, and the accumulation of compatible solutes, were also observed in S. algae. In particular, genes involved in peptidoglycan biosynthesis and DNA repair were highly and steadily up-regulated, accompanied by rapid and instantaneous enhancement of the transcription of large- and small-ribosome subunits, which suggested that the structural changes in the cell wall and some stressful responses occurred in S. algae. Furthermore, the transcription of genes involved in the tricarboxylic acid (TCA) cycle and the glycolytic pathway was decreased, whereas the transcription of genes involved in anaerobic respiration was increased. These results, demonstrating the multi-pathway reactions of S. algae in response to salt stress, increase our understanding of the microbial stress response mechanisms.
Conflict of interest statement
Figures


References
-
- Verma P, Pandey PK, Gupta AK, Kim HJ, Baik KS, et al. (2011) Shewanella indica sp. nov., isolated from sediment of the Arabian Sea. Int J Syst Evol Microbiol 61: 2058–2064. - PubMed
-
- Nath R, Saikia L, Choudhury G, Das PP (2011) Isolation of Shewanella algae from rectal swabs of patients with bloody diarrhoea. Indian J Med Microbiol 29: 422–425. - PubMed
-
- Zong Z (2011) Nosocomial peripancreatic infection associated with Shewanella xiamenensis. J Med Microbiol 60: 1387–1390. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

