Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli
- PMID: 24789184
- PMCID: PMC4097710
- DOI: 10.1128/JCM.00313-14
Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli
Abstract
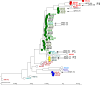
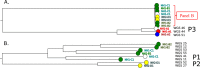
Next-generation sequencing (NGS) has the potential to provide typing results and detect resistance genes in a single assay, thus guiding timely treatment decisions and allowing rapid tracking of transmission of resistant clones. We evaluated the performance of a new NGS assay (Hospital Acquired Infection BioDetection System; Pathogenica) during an outbreak of sequence type 131 (ST131) Escherichia coli infections in a nursing home in The Netherlands. The assay was performed on 56 extended-spectrum-beta-lactamase (ESBL) E. coli isolates collected during 2 prevalence surveys (March and May 2013). Typing results were compared to those of amplified fragment length polymorphism (AFLP), whereby we visually assessed the agreement of the BioDetection phylogenetic tree with clusters defined by AFLP. A microarray was considered the gold standard for detection of resistance genes. AFLP identified a large cluster of 31 indistinguishable isolates on adjacent departments, indicating clonal spread. The BioDetection phylogenetic tree showed that all isolates of this outbreak cluster were strongly related, while the further arrangement of the tree also largely agreed with other clusters defined by AFLP. The BioDetection assay detected ESBL genes in all but 1 isolate (sensitivity, 98%) but was unable to discriminate between ESBL and non-ESBL TEM and SHV beta-lactamases or to specify CTX-M genes by group. The performance of the hospital-acquired infection (HAI) BioDetection System for typing of E. coli isolates compared well with the results of AFLP. Its performance with larger collections from different locations, and for typing of other species, was not evaluated and needs further study.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawekey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 17:1216–1223. 10.3201/eid1707.110209 - DOI - PMC - PubMed
-
- Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13:19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

