Improved molecular typing assay for rhinovirus species A, B, and C
- PMID: 24789198
- PMCID: PMC4097758
- DOI: 10.1128/JCM.00075-14
Improved molecular typing assay for rhinovirus species A, B, and C
Abstract
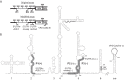
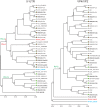
Human rhinoviruses (RVs), comprising three species (A, B, and C) of the genus Enterovirus, are responsible for the majority of upper respiratory tract infections and are associated with severe lower respiratory tract illnesses such as pneumonia and asthma exacerbations. High genetic diversity and continuous identification of new types necessitate regular updating of the diagnostic assays for the accurate and comprehensive detection of circulating RVs. Methods for molecular typing based on phylogenetic comparisons of a variable fragment in the 5' untranslated region were improved to increase assay sensitivity and to eliminate nonspecific amplification of human sequences, which are observed occasionally in clinical samples. A modified set of primers based on new sequence information and improved buffers and enzymes for seminested PCR assays provided higher specificity and sensitivity for virus detection. In addition, new diagnostic primers were designed for unequivocal species and type assignments for RV-C isolates, based on phylogenetic analysis of partial VP4/VP2 coding sequences. The improved assay was evaluated by typing RVs in >3,800 clinical samples. RVs were successfully detected and typed in 99% of the samples that were RV positive in multiplex diagnostic assays.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, Le Souef PN. 2011. Association between human rhinovirus C and severity of acute asthma in children. Eur. Respir. J. 37:1037–1042. 10.1183/09031936.00092410 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

