Mechanistic differences between nucleic acid chaperone activities of the Gag proteins of Rous sarcoma virus and human immunodeficiency virus type 1 are attributed to the MA domain
- PMID: 24789780
- PMCID: PMC4097784
- DOI: 10.1128/JVI.00736-14
Mechanistic differences between nucleic acid chaperone activities of the Gag proteins of Rous sarcoma virus and human immunodeficiency virus type 1 are attributed to the MA domain
Abstract
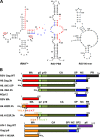
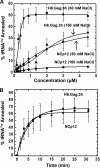
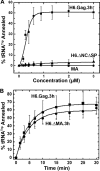
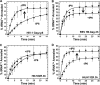
Host cell tRNAs are recruited for use as primers to initiate reverse transcription in retroviruses. Human immunodeficiency virus type 1 (HIV-1) uses tRNA(Lys3) as the replication primer, whereas Rous sarcoma virus (RSV) uses tRNA(Trp). The nucleic acid (NA) chaperone function of the nucleocapsid (NC) domain of HIV-1 Gag is responsible for annealing tRNA(Lys3) to the genomic RNA (gRNA) primer binding site (PBS). Compared to HIV-1, little is known about the chaperone activity of RSV Gag. In this work, using purified RSV Gag containing an N-terminal His tag and a deletion of the majority of the protease domain (H6.Gag.3h), gel shift assays were used to monitor the annealing of tRNA(Trp) to a PBS-containing RSV RNA. Here, we show that similar to HIV-1 Gag lacking the p6 domain (GagΔp6), RSV H6.Gag.3h is a more efficient chaperone on a molar basis than NC; however, in contrast to the HIV-1 system, both RSV H6.Gag.3h and NC have comparable annealing rates at protein saturation. The NC domain of RSV H6.Gag.3h is required for annealing, whereas deletion of the matrix (MA) domain, which stimulates the rate of HIV-1 GagΔp6 annealing, has little effect on RSV H6.Gag.3h chaperone function. Competition assays confirmed that RSV MA binds inositol phosphates (IPs), but in contrast to HIV-1 GagΔp6, IPs do not stimulate RSV H6.Gag.3h chaperone activity unless the MA domain is replaced with HIV-1 MA. We conclude that differences in the MA domains are primarily responsible for mechanistic differences in RSV and HIV-1 Gag NA chaperone function. Importance: Mounting evidence suggests that the Gag polyprotein is responsible for annealing primer tRNAs to the PBS to initiate reverse transcription in retroviruses, but only HIV-1 Gag chaperone activity has been demonstrated in vitro. Understanding RSV Gag's NA chaperone function will allow us to determine whether there is a common mechanism among retroviruses. This report shows for the first time that full-length RSV Gag lacking the protease domain is a highly efficient NA chaperone in vitro, and NC is required for this activity. In contrast to results obtained for HIV-1 Gag, due to the weak nucleic acid binding affinity of the RSV MA domain, inositol phosphates do not regulate RSV Gag-facilitated tRNA annealing despite the fact that they bind to MA. These studies provide insight into the viral regulation of tRNA primer annealing, which is a potential target for antiretroviral therapy.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
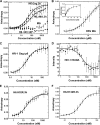
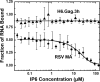
Similar articles
-
Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding.J Virol. 2011 Feb;85(4):1594-603. doi: 10.1128/JVI.01809-10. Epub 2010 Dec 1. J Virol. 2011. PMID: 21123373 Free PMC article.
-
The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag.Retrovirology. 2016 Mar 17;13:18. doi: 10.1186/s12977-016-0245-1. Retrovirology. 2016. PMID: 26987314 Free PMC article.
-
The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site.J Virol. 1999 May;73(5):4251-6. doi: 10.1128/JVI.73.5.4251-4256.1999. J Virol. 1999. PMID: 10196321 Free PMC article.
-
Aspects of HIV-1 assembly that promote primer tRNA(Lys3) annealing to viral RNA.Virus Res. 2012 Nov;169(2):340-8. doi: 10.1016/j.virusres.2012.06.001. Epub 2012 Jun 12. Virus Res. 2012. PMID: 22698876 Review.
-
Nucleic acid chaperone activity of retroviral Gag proteins.RNA Biol. 2010 Nov-Dec;7(6):700-5. doi: 10.4161/rna.7.6.13685. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045546 Free PMC article. Review.
Cited by
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. PMID: 38293010 Free PMC article. Updated. Preprint.
-
RNA-Binding Domains of Heterologous Viral Proteins Substituted for Basic Residues in the RSV Gag NC Domain Restore Specific Packaging of Genomic RNA.Viruses. 2020 Mar 27;12(4):370. doi: 10.3390/v12040370. Viruses. 2020. PMID: 32230826 Free PMC article.
-
HIV-1 Pr55Gag binds genomic and spliced RNAs with different affinity and stoichiometry.RNA Biol. 2017 Jan 2;14(1):90-103. doi: 10.1080/15476286.2016.1256533. Epub 2016 Nov 14. RNA Biol. 2017. PMID: 27841704 Free PMC article.
-
Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag.Viruses. 2016 Sep 19;8(9):256. doi: 10.3390/v8090256. Viruses. 2016. PMID: 27657107 Free PMC article.
-
Rous Sarcoma Virus Genomic RNA Dimerization Capability In Vitro Is Not a Prerequisite for Viral Infectivity.Viruses. 2020 May 22;12(5):568. doi: 10.3390/v12050568. Viruses. 2020. PMID: 32455905 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

