Activation of GATA4 gene expression at the early stage of cardiac specification
- PMID: 24790981
- PMCID: PMC3982529
- DOI: 10.3389/fchem.2014.00012
Activation of GATA4 gene expression at the early stage of cardiac specification
Abstract
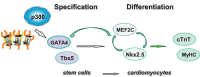
Currently, there are no effective treatments to directly repair damaged heart tissue after cardiac injury since existing therapies focus on rescuing or preserving reversibly damaged tissue. Cell-based therapies using cardiomyocytes generated from stem cells present a promising therapeutic approach to directly replace damaged myocardium with new healthy tissue. However, the molecular mechanisms underlying the commitment of stem cells into cardiomyocytes are not fully understood and will be critical to guide this new technology into the clinic. Since GATA4 is a critical regulator of cardiac differentiation, we examined the molecular basis underlying the early activation of GATA4 gene expression during cardiac differentiation of pluripotent stem cells. Our studies demonstrate the direct involvement of histone acetylation and transcriptional coactivator p300 in the regulation of GATA4 gene expression. More importantly, we show that histone acetyltransferase (HAT) activity is important for GATA4 gene expression with the use of curcumin, a HAT inhibitor. In addition, the widely used histone deacetylase inhibitor valproic acid enhances both histone acetylation and cardiac specification.
Keywords: GATA4; cardiac differentiation; cardiomyocytes; gene regulation; histone acetylation.
Figures
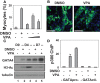

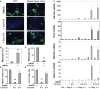
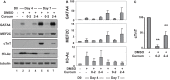
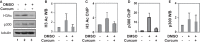
References
-
- Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U., et al. (2004). Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163–51171 10.1074/jbc.M409024200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

