A family of null models to distinguish between environmental filtering and biotic interactions in functional diversity patterns
- PMID: 24791143
- PMCID: PMC4003529
- DOI: 10.1111/jvs.12031
A family of null models to distinguish between environmental filtering and biotic interactions in functional diversity patterns
Abstract
Questions: Traditional null models used to reveal assembly processes from functional diversity patterns are not tailored for comparing different spatial and evolutionary scales. In this study, we present and explore a family of null models that can help disentangling assembly processes at their appropriate scales and thereby elucidate the ecological drivers of community assembly.
Location: French Alps.
Methods: Our approach gradually constrains null models by: (1) filtering out species not able to survive in the regional conditions in order to reduce the spatial scale, and (2) shuffling species only within lineages of different ages to reduce the evolutionary scale of the analysis. We first tested and validated this approach using simulated communities. We then applied it to study the functional diversity patterns of the leaf-height-seed strategy of plant communities in the French Alps.
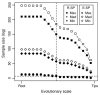
Results: Using simulations, we found that reducing the spatial scale correctly detected a signature of competition (functional divergence) even when environmental filtering produced an overlaying signal of functional convergence. However, constraining the evolutionary scale did not change the identified functional diversity patterns. In the case study of alpine plant communities, investigating scale effects revealed that environmental filtering had a strong influence at larger spatial and evolutionary scales and that neutral processes were more important at smaller scales. In contrast to the simulation study results, decreasing the evolutionary scale tended to increase patterns of functional divergence.
Conclusion: We argue that the traditional null model approach can only identify a single main process at a time and suggest to rather use a family of null models to disentangle intertwined assembly processes acting across spatial and evolutionary scales.
Keywords: Assembly rules; Biotic and abiotic filtering; Limiting similarity; Null models; Simulated communities.
Figures


References
-
- Albert CH, Thuiller W, Yoccoz NG, Douzet R, Aubert S, Lavorel S. A multi-trait approach reveals the structure and the relative importance of intravs. interspecific variability in plant traits. Functional Ecology. 2010;24:1192–1201.
-
- Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. - PubMed
-
- Boulangeat I, Lavergne S, Van Es J, Garraud L, Thuiller W. Niche breadth, rarity and ecological characteristics within a regional flora spanning large environmental gradients. Journal of Biogeography. 2012b;39:204–214.
-
- Carboni M, Acosta A, Ricotta C. Differences in functional diversity among plant communities on Mediterranean coastal dunes are driven by their phylogenetic history. Journal of Vegetation Science. 2013;24:932–941.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
