A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy
- PMID: 24792170
- PMCID: PMC4041457
- DOI: 10.1093/nar/gku284
A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy
Abstract
DNA-damage response machinery is crucial to maintain the genomic integrity of cells, by enabling effective repair of even highly lethal lesions such as DNA double-strand breaks (DSBs). Defects in specific genes acquired through mutations, copy-number alterations or epigenetic changes can alter the balance of these pathways, triggering cancerous potential in cells. Selective killing of cancer cells by sensitizing them to further DNA damage, especially by induction of DSBs, therefore requires careful modulation of DSB-repair pathways. Here, we review the latest knowledge on the two DSB-repair pathways, homologous recombination and non-homologous end joining in human, describing in detail the functions of their components and the key mechanisms contributing to the repair. Such an in-depth characterization of these pathways enables a more mechanistic understanding of how cells respond to therapies, and suggests molecules and processes that can be explored as potential therapeutic targets. One such avenue that has shown immense promise is via the exploitation of synthetic lethal relationships, for which the BRCA1-PARP1 relationship is particularly notable. Here, we describe how this relationship functions and the manner in which cancer cells acquire therapy resistance by restoring their DSB repair potential.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
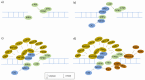

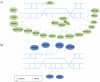
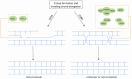
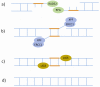
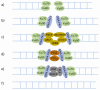
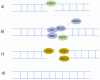
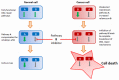
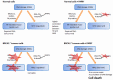
References
-
- Khanna K.K., Jackson S.P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. - PubMed
-
- Rich T., Allen R.L., Wyllie A.H. Defying death after DNA damage. Nature. 2000;407:777–783. - PubMed
-
- Fojo T. Cancer, DNA repair mechanisms, and resistance to chemotherapy. J. Natl. Cancer Inst. 2001;93:1434–1436. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

