Creating stable stem regions for loop elongation in Fcabs - insights from combining yeast surface display, in silico loop reconstruction and molecular dynamics simulations
- PMID: 24792385
- PMCID: PMC4118681
- DOI: 10.1016/j.bbapap.2014.04.020
Creating stable stem regions for loop elongation in Fcabs - insights from combining yeast surface display, in silico loop reconstruction and molecular dynamics simulations
Abstract
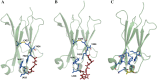
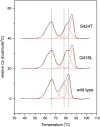
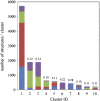
Fcabs (Fc antigen binding) are crystallizable fragments of IgG where the C-terminal structural loops of the CH3 domain are engineered for antigen binding. For the design of libraries it is beneficial to know positions that will permit loop elongation to increase the potential interaction surface with antigen. However, the insertion of additional loop residues might impair the immunoglobulin fold. In the present work we have probed whether stabilizing mutations flanking the randomized and elongated loop region improve the quality of Fcab libraries. In detail, 13 libraries were constructed having the C-terminal part of the EF loop randomized and carrying additional residues (1, 2, 3, 5 or 10, respectively) in the absence and presence of two flanking mutations. The latter have been demonstrated to increase the thermal stability of the CH3 domain of the respective solubly expressed proteins. Assessment of the stability of the libraries expressed on the surface of yeast cells by flow cytometry demonstrated that loop elongation was considerably better tolerated in the stabilized libraries. By using in silico loop reconstruction and mimicking randomization together with MD simulations the underlying molecular dynamics were investigated. In the presence of stabilizing stem residues the backbone flexibility of the engineered EF loop as well as the fluctuation between its accessible conformations were decreased. In addition the CD loop (but not the AB loop) and most of the framework regions were rigidified. The obtained data are discussed with respect to the design of Fcabs and available data on the relation between flexibility and affinity of CDR loops in Ig-like molecules.
Keywords: Fcab; Loop reconstruction; Molecular dynamics simulation; Protein engineering; Therapeutic antibody fragment; Yeast surface display.
Copyright © 2014. Published by Elsevier B.V.
Figures


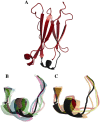
References
-
- Perutz M.F. Introduction to protein-structure — Branden,C, Tooze. J. Nat. 1991;353:311.
-
- Halaby D.M., Poupon A., Mornon J.P. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563–571. - PubMed
-
- Traxlmayr M.W., Wozniak-Knopp G., Antes B., Stadlmayr G., Ruker F., Obinger C. Integrin binding human antibody constant domains-probing the C-terminal structural loops for grafting the RGD motif. J. Biotechnol. 2011;155:193–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

