Connective tissue growth factor is a target of notch signaling in cells of the osteoblastic lineage
- PMID: 24792956
- PMCID: PMC4069863
- DOI: 10.1016/j.bone.2014.04.028
Connective tissue growth factor is a target of notch signaling in cells of the osteoblastic lineage
Abstract
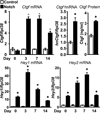
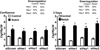
Connective tissue growth factor (Ctgf) or CCN2 is a protein synthesized by osteoblasts necessary for skeletal homeostasis, although its overexpression inhibits osteogenic signals and bone formation. Ctgf is induced by bone morphogenetic proteins, transforming growth factor β and Wnt; and in the present studies, we explored whether Notch regulated Ctgf expression in osteoblasts. We employed Rosa(Notch) mice, where the Notch intracellular domain (NICD) is expressed following the excision of a STOP cassette, placed between the Rosa26 promoter and NICD. Notch was activated by transduction of adenoviral vectors expressing Cre recombinase (Ad-CMV-Cre). Notch induced Ctgf mRNA levels in a time dependent manner and increased Ctgf heterogeneous nuclear RNA. Notch also destabilized Ctgf mRNA shortening its half-life from 13h to 3h. The effect of Notch on Ctgf expression was lost following Rbpjκ downregulation, demonstrating that it was mediated by Notch canonical signaling. However, downregulation of the classic Notch target genes Hes1, Hey1 and Hey2 did not modify the effect of Notch on Ctgf expression. Wild type osteoblasts exposed to immobilized Delta-like 1 displayed enhanced Notch signaling and increased Ctgf expression. In addition to the effects of Notch in vitro, Notch induced Ctgf in vivo, and calvariae and femurs from Rosa(Notch) mice mated with transgenics expressing the Cre recombinase in cells of the osteoblastic lineage exhibited increased expression of Ctgf. In conclusion, Ctgf is a target of Notch canonical signaling in osteoblasts, and may act in concert with Notch to regulate skeletal homeostasis.
Keywords: CCN proteins; Notch; connective tissue growth factor; osteoblasts; transcription.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




References
-
- Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. - PubMed
-
- Gazzerro E, Canalis E. Skeletal actions of insulin-like growth factors. Expert Rev Endocrinol Metab. 2006;1:47–56. - PubMed
-
- Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013;9:575–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

