Tissue-specific signals control reversible program of localization and functional polarization of macrophages
- PMID: 24792964
- PMCID: PMC4137874
- DOI: 10.1016/j.cell.2014.04.016
Tissue-specific signals control reversible program of localization and functional polarization of macrophages
Abstract
Tissue-resident macrophages are highly heterogeneous in terms of their functions and phenotypes as a consequence of adaptation to different tissue environments. Local tissue-derived signals are thought to control functional polarization of resident macrophages; however, the identity of these signals remains largely unknown. It is also unknown whether functional heterogeneity is a result of irreversible lineage-specific differentiation or a consequence of continuous but reversible induction of diverse functional programs. Here, we identified retinoic acid as a signal that induces tissue-specific localization and functional polarization of peritoneal macrophages through the reversible induction of transcription factor GATA6. We further found that GATA6 in macrophages regulates gut IgA production through peritoneal B-1 cells. These results provide insight into the regulation of tissue-resident macrophage functional specialization by tissue-derived signals.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
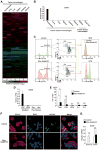
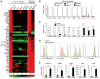
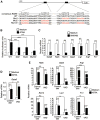




Comment in
-
Macrophages: Peritoneal population depends on GATA6.Nat Rev Immunol. 2014 Jun;14(6):360. doi: 10.1038/nri3695. Nat Rev Immunol. 2014. PMID: 24854587 No abstract available.
References
-
- Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. - PubMed
-
- Barth MW, Hendrzak JA, Melnicoff MJ, Morahan PS. Review of the macrophage disappearance reaction. J Leukoc Biol. 1995;57:361–367. - PubMed
-
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. - PubMed
-
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

