Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins
- PMID: 24793022
- PMCID: PMC4052017
- DOI: 10.1007/s11103-014-0194-9
Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins
Abstract
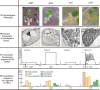
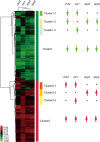
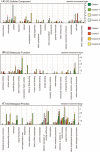
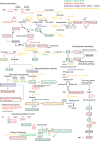
We used four mutants having albino or pale green phenotypes with disrupted nuclear-encoded chloroplast proteins to analyze the regulatory system of metabolites in chloroplast. We performed an integrated analyses of transcriptomes and metabolomes of the four mutants. Transcriptome analysis was carried out using the Agilent Arabidopsis 2 Oligo Microarray, and metabolome analysis with two mass spectrometers; a direct-infusion Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR/MS) and a gas chromatograph-time of flight mass spectrometer. Among approximately 200 known metabolites detected by the FT-ICR/MS, 71 metabolites showed significant changes in the mutants when compared with controls (Ds donor plants). Significant accumulation of several amino acids (glutamine, glutamate and asparagine) was observed in the albino and pale green mutants. Transcriptome analysis revealed altered expressions of genes in several metabolic pathways. For example, genes involved in the tricarboxylic acid cycle, the oxidative pentose phosphate pathway, and the de novo purine nucleotide biosynthetic pathway were up-regulated. These results suggest that nitrogen assimilation is constitutively promoted in the albino and pale green mutants. The accumulation of ammonium ions in the albino and pale green mutants was consistently higher than in Ds donor lines. Furthermore, genes related to pyridoxin accumulation and the de novo purine nucleotide biosynthetic pathway were up-regulated, which may have occurred as a result of the accumulation of glutamine in the albino and pale green mutants. The difference in metabolic profiles seems to be correlated with the disruption of chloroplast internal membrane structures in the mutants. In albino mutants, the alteration of metabolites accumulation and genes expression is stronger than pale green mutants.
Figures

References
-
- Booij-James IS, Dube SK, Jansen MA, Edelman M, Mattoo AK. Ultraviolet-B radiation impacts light-mediated turnover of the photosystem II reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiol. 2000;124:1275–1284. doi: 10.1104/pp.124.3.1275. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

