Sex-biased gene expression and evolution of the x chromosome in nematodes
- PMID: 24793291
- PMCID: PMC4096367
- DOI: 10.1534/genetics.114.163311
Sex-biased gene expression and evolution of the x chromosome in nematodes
Abstract
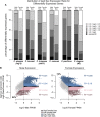
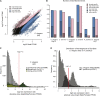
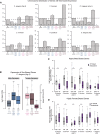
Studies of X chromosome evolution in various organisms have indicated that sex-biased genes are nonrandomly distributed between the X and autosomes. Here, to extend these studies to nematodes, we annotated and analyzed X chromosome gene content in four Caenorhabditis species and in Pristionchus pacificus. Our gene expression analyses comparing young adult male and female mRNA-seq data indicate that, in general, nematode X chromosomes are enriched for genes with high female-biased expression and depleted of genes with high male-biased expression. Genes with low sex-biased expression do not show the same trend of X chromosome enrichment and depletion. Combined with the observation that highly sex-biased genes are primarily expressed in the gonad, differential distribution of sex-biased genes reflects differences in evolutionary pressures linked to tissue-specific regulation of X chromosome transcription. Our data also indicate that X dosage imbalance between males (XO) and females (XX) is influential in shaping both expression and gene content of the X chromosome. Predicted upregulation of the single male X to match autosomal transcription (Ohno's hypothesis) is supported by our observation that overall transcript levels from the X and autosomes are similar for highly expressed genes. However, comparison of differentially located one-to-one orthologs between C. elegans and P. pacificus indicates lower expression of X-linked orthologs, arguing against X upregulation. These contradicting observations may be reconciled if X upregulation is not a global mechanism but instead acts locally on a subset of tissues and X-linked genes that are dosage sensitive.
Keywords: X chromosome evolution; dosage compensation; genomics; nematode; sex-biased gene expression.
Copyright © 2014 by the Genetics Society of America.
Figures





References
-
- Arnqvist G., 2004. Sexual conflict and sexual selection: lost in the chase. Evolution 58: 1383–1393. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

