Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance
- PMID: 24793693
- PMCID: PMC4267251
- DOI: 10.1016/j.molcel.2014.03.045
Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance
Abstract
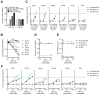
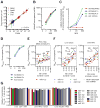
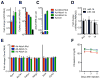
Recent studies have reported that competitive endogenous RNAs (ceRNAs) can act as sponges for a microRNA (miRNA) through their binding sites and that changes in ceRNA abundances from individual genes can modulate the activity of miRNAs. Consideration of this hypothesis would benefit from knowing the quantitative relationship between a miRNA and its endogenous target sites. Here, we altered intracellular target site abundance through expression of an miR-122 target in hepatocytes and livers and analyzed the effects on miR-122 target genes. Target repression was released in a threshold-like manner at high target site abundance (≥1.5 × 10(5) added target sites per cell), and this threshold was insensitive to the effective levels of the miRNA. Furthermore, in response to extreme metabolic liver disease models, global target site abundance of hepatocytes did not change sufficiently to affect miRNA-mediated repression. Thus, modulation of miRNA target abundance is unlikely to cause significant effects on gene expression and metabolism through a ceRNA effect.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Competitive endogenous RNAs cannot alter microRNA function in vivo.Mol Cell. 2014 Jun 5;54(5):711-3. doi: 10.1016/j.molcel.2014.05.023. Mol Cell. 2014. PMID: 24905003
Similar articles
-
Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression.Mol Cell. 2016 Nov 3;64(3):565-579. doi: 10.1016/j.molcel.2016.09.027. Epub 2016 Oct 27. Mol Cell. 2016. PMID: 27871486 Free PMC article.
-
Endogenous microRNA sponges: evidence and controversy.Nat Rev Genet. 2016 May;17(5):272-83. doi: 10.1038/nrg.2016.20. Epub 2016 Apr 4. Nat Rev Genet. 2016. PMID: 27040487 Review.
-
Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer.PLoS One. 2017 Feb 10;12(2):e0171661. doi: 10.1371/journal.pone.0171661. eCollection 2017. PLoS One. 2017. PMID: 28187158 Free PMC article.
-
Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk.Crit Rev Biochem Mol Biol. 2018 Jun;53(3):231-245. doi: 10.1080/10409238.2018.1447542. Epub 2018 Mar 23. Crit Rev Biochem Mol Biol. 2018. PMID: 29569941 Free PMC article. Review.
-
Discovering and Constructing ceRNA-miRNA-Target Gene Regulatory Networks during Anther Development in Maize.Int J Mol Sci. 2019 Jul 15;20(14):3480. doi: 10.3390/ijms20143480. Int J Mol Sci. 2019. PMID: 31311189 Free PMC article.
Cited by
-
Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria.Nat Rev Microbiol. 2016 Dec;14(12):775-784. doi: 10.1038/nrmicro.2016.129. Epub 2016 Sep 19. Nat Rev Microbiol. 2016. PMID: 27640758 Review.
-
Long Non-Coding RNAs in Liver Cancer and Nonalcoholic Steatohepatitis.Noncoding RNA. 2020 Aug 29;6(3):34. doi: 10.3390/ncrna6030034. Noncoding RNA. 2020. PMID: 32872482 Free PMC article. Review.
-
Plant bioactive compounds driven microRNAs (miRNAs): A potential source and novel strategy targeting gene and cancer therapeutics.Noncoding RNA Res. 2024 Jun 8;9(4):1140-1158. doi: 10.1016/j.ncrna.2024.06.003. eCollection 2024 Dec. Noncoding RNA Res. 2024. PMID: 39022680 Free PMC article.
-
A network-biology perspective of microRNA function and dysfunction in cancer.Nat Rev Genet. 2016 Dec;17(12):719-732. doi: 10.1038/nrg.2016.134. Epub 2016 Oct 31. Nat Rev Genet. 2016. PMID: 27795564 Review.
-
Past, present, and future of circRNAs.EMBO J. 2019 Aug 15;38(16):e100836. doi: 10.15252/embj.2018100836. Epub 2019 Jul 25. EMBO J. 2019. PMID: 31343080 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

