Synthetic analog and digital circuits for cellular computation and memory
- PMID: 24794536
- PMCID: PMC4237220
- DOI: 10.1016/j.copbio.2014.04.009
Synthetic analog and digital circuits for cellular computation and memory
Abstract
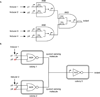
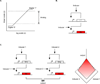
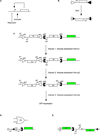
Biological computation is a major area of focus in synthetic biology because it has the potential to enable a wide range of applications. Synthetic biologists have applied engineering concepts to biological systems in order to construct progressively more complex gene circuits capable of processing information in living cells. Here, we review the current state of computational genetic circuits and describe artificial gene circuits that perform digital and analog computation. We then discuss recent progress in designing gene networks that exhibit memory, and how memory and computation have been integrated to yield more complex systems that can both process and record information. Finally, we suggest new directions for engineering biological circuits capable of computation.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 2012;13:455–468. - PubMed
-
- Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J. Biotechnol. 2007;130:329–345. - PubMed
-
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial Synthesis of Genetic Networks. Science. 2002;296:1466–1470. - PubMed
-
- Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

