Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors
- PMID: 24794633
- PMCID: PMC4045103
- DOI: 10.1016/j.devcel.2014.04.001
Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors
Abstract
In vertebrates, pluripotent pharyngeal mesoderm progenitors produce the cardiac precursors of the second heart field as well as the branchiomeric head muscles and associated stem cells. However, the mechanisms underlying the transition from multipotent progenitors to distinct muscle precursors remain obscured by the complexity of vertebrate embryos. Using Ciona intestinalis as a simple chordate model, we show that bipotent cardiopharyngeal progenitors are primed to activate both heart and pharyngeal muscle transcriptional programs, which progressively become restricted to corresponding precursors. The transcription factor COE (Collier/OLF/EBF) orchestrates the transition to pharyngeal muscle fate both by promoting an MRF-associated myogenic program in myoblasts and by maintaining an undifferentiated state in their sister cells through Notch-mediated lateral inhibition. The latter are stem cell-like muscle precursors that form most of the juvenile pharyngeal muscles. We discuss the implications of our findings for the development and evolution of the chordate cardiopharyngeal mesoderm.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
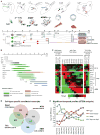
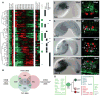

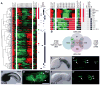
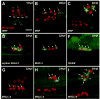
References
-
- Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. - PubMed
-
- Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Current opinion in genetics & development. 2009;19:444–453. - PubMed
-
- Chiba S, Sasaki A, Nakayama A, Takamura K, Satoh N. Development of Ciona intestinalis juveniles (through 2nd ascidian stage) Zoological science. 2004;21:285–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

