α1-adrenergic receptor signaling in osteoblasts regulates clock genes and bone morphogenetic protein 4 expression through up-regulation of the transcriptional factor nuclear factor IL-3 (Nfil3)/E4 promoter-binding protein 4 (E4BP4)
- PMID: 24794868
- PMCID: PMC4059158
- DOI: 10.1074/jbc.M113.546135
α1-adrenergic receptor signaling in osteoblasts regulates clock genes and bone morphogenetic protein 4 expression through up-regulation of the transcriptional factor nuclear factor IL-3 (Nfil3)/E4 promoter-binding protein 4 (E4BP4)
Abstract
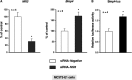
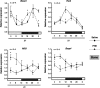
Several studies have demonstrated that the α1-adrenergic receptor (AR) plays an important role in regulating cell growth and function in osteoblasts. However, the physiological role of α1-AR signaling in bone metabolism is largely unknown. In this study, the stimulation of phenylephrine (PHE), a nonspecific α1-AR agonist, increased the transcriptional factor Nfil3/E4BP4 and led to the rhythmic expression of bone morphogenetic protein 4 (Bmp4) in MC3T3-E1 osteoblastic cells. We also showed that Bmp4 mRNA expression peaked in bone near zeitgeber time 8 in a 24-h rhythm. Furthermore, the expression of Nfil3 and Bmp4 displayed a circadian pattern with opposing phases, which suggested that Nfil3 repressed the expression of the Bmp4 gene during a circadian cycle. On a molecular level, both loss-of-function and gain-of-function experiments demonstrated that Nfil3/E4BP4 negatively regulated Bmp4 expression in osteoblasts. Furthermore, the systemic administration of PHE increased the expression of Nfil3 mRNA in bone, whereas it decreased that of Bmp4 mRNA. The expression of Bmp4 mRNA was decreased significantly by exposure to PHE, and this was concomitant with the increase in Nfil3 binding to the D-box-containing Bmp4 promoter region in MC3T3-E1 cells, which indicates that the expression of Nfil3 by α1-AR signaling can bind directly to the Bmp4 promoter and inhibit Bmp4 expression in osteoblasts. Our results suggest that α1-AR signaling regulates clock genes and Bmp4 expression in osteoblasts. Moreover, α1-AR signaling negatively regulated Bmp4 expression by up-regulating the transcriptional factor Nfil3/E4BP4 in osteoblasts.
Keywords: Adrenergic Receptor; Bmp4; Bone Morphogenetic Protein (BMP); Clock Gene; Nfil3; Osteoblast; Transcription Factor; α1-Adrenergic Receptor.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








References
-
- Prinster S. C., Hague C, Hall R. A. (2005) Heterodimerization of G protein-coupled receptors: specificity and functional significance. Pharmacol. Rev. 7, 289–298 - PubMed
-
- Philipp M., Hein L. (2004) Adrenergic receptor knockout mice: distinct functions of 9 receptor subtypes. Pharmacol. Ther. 101, 65–74 - PubMed
-
- Wettschureck N., Offermanns S. (2005) Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 - PubMed
-
- Takeda S., Elefteriou F., Levasseur R., Liu X., Zhao L., Parker K. L., Armstrong D., Ducy P., Karsenty G. (2002) Leptin regulates bone formation via the sympathetic nervous system. Cell 111, 305–317 - PubMed
-
- Togari A., Arai M., Kondo A. (2005) The role of the sympathetic nervous system in controlling bone metabolism. Expert Opin. Ther. Targets 9, 931–940 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

