The available SRL3 deletion strain of Saccharomyces cerevisiae contains a truncation of DNA damage tolerance protein Mms2: Implications for Srl3 and Mms2 functions
- PMID: 24795789
- PMCID: PMC4008323
- DOI: 10.5580/42c
The available SRL3 deletion strain of Saccharomyces cerevisiae contains a truncation of DNA damage tolerance protein Mms2: Implications for Srl3 and Mms2 functions
Abstract
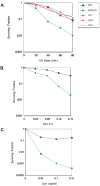
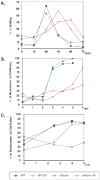
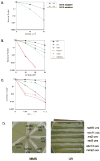
A screen of the commercially available collection of haploid deletion mutants of Saccharomyces cerevisiae for spontaneous mutator mutants newly identified a deletion of SRL3. This gene had been previously isolated as a suppressor of lethality of checkpoint kinase deletions if overexpressed. We found DNA damage sensitivity and extended checkpoint arrests to be associated with this strain. However, when crossed to wild-type, a mutant gene conferring these phenotypes was found to segregate from the SRL3 deletion. The mutation was identified as a C-terminal truncation of Mms2, an E2 ubiquitin conjugating enzyme involved in error-free replicative bypass of lesions. This confirmed an earlier report that Mms2 may be required to restrain error-prone polymerase ζ activity and underscored that residues of the C-terminus are necessary for Mms2 function. Srl3, on the other hand, does not appear to influence DNA damage sensitivity or spontaneous mutability if deleted. However, the absence of these phenotypes does not contradict its likely role as a positive regulator of dNTP levels.
Keywords: Checkpoints; DNA Damage Tolerance; DNA Repair; Mutagenesis; Yeast; dNTP Pools.
Figures

Similar articles
-
MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway.Proc Natl Acad Sci U S A. 1998 May 12;95(10):5678-83. doi: 10.1073/pnas.95.10.5678. Proc Natl Acad Sci U S A. 1998. PMID: 9576943 Free PMC article.
-
A novel role for Mms2 in the control of spontaneous mutagenesis and Pol3 abundance.DNA Repair (Amst). 2023 May;125:103484. doi: 10.1016/j.dnarep.2023.103484. Epub 2023 Mar 13. DNA Repair (Amst). 2023. PMID: 36934633
-
Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae.Mutat Res. 1999 Sep 13;435(1):1-11. doi: 10.1016/s0921-8777(99)00034-8. Mutat Res. 1999. PMID: 10526212
-
Role of PSO genes in repair of DNA damage of Saccharomyces cerevisiae.Mutat Res. 2003 Nov;544(2-3):179-93. doi: 10.1016/j.mrrev.2003.06.018. Mutat Res. 2003. PMID: 14644320 Review.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
References
-
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2005. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005.
-
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. - PubMed
-
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. - PubMed
-
- Brendel M, Haynes RH. Interactions among genes controlling sensitivity to radiation and alkylation in yeast. Mol Gen Genet. 1973;125:197–216. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
