Functional characterization of the Xanthophyllomyces dendrorhous farnesyl pyrophosphate synthase and geranylgeranyl pyrophosphate synthase encoding genes that are involved in the synthesis of isoprenoid precursors
- PMID: 24796858
- PMCID: PMC4010515
- DOI: 10.1371/journal.pone.0096626
Functional characterization of the Xanthophyllomyces dendrorhous farnesyl pyrophosphate synthase and geranylgeranyl pyrophosphate synthase encoding genes that are involved in the synthesis of isoprenoid precursors
Abstract
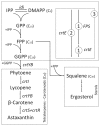
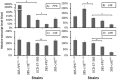
The yeast Xanthophyllomyces dendrorhous synthesizes the carotenoid astaxanthin, which has applications in biotechnology because of its antioxidant and pigmentation properties. However, wild-type strains produce too low amounts of carotenoids to be industrially competitive. Considering this background, it is indispensable to understand how the synthesis of astaxanthin is controlled and regulated in this yeast. In this work, the steps leading to the synthesis of the carotenoid precursor geranylgeranyl pyrophosphate (GGPP, C20) in X. dendrorhous from isopentenyl pyrophosphate (IPP, C5) and dimethylallyl pyrophosphate (DMAPP, C5) was characterized. Two prenyl transferase encoding genes, FPS and crtE, were expressed in E. coli. The enzymatic assays using recombinant E. coli protein extracts demonstrated that FPS and crtE encode a farnesyl pyrophosphate (FPP, C15) synthase and a GGPP-synthase, respectively. X. dendrorhous FPP-synthase produces geranyl pyrophosphate (GPP, C10) from IPP and DMAPP and FPP from IPP and GPP, while the X. dendrorhous GGPP-synthase utilizes only FPP and IPP as substrates to produce GGPP. Additionally, the FPS and crtE genes were over-expressed in X. dendrorhous, resulting in an increase of the total carotenoid production. Because the parental strain is diploid, the deletion of one of the alleles of these genes did not affect the total carotenoid production, but the composition was significantly altered. These results suggest that the over-expression of these genes might provoke a higher carbon flux towards carotenogenesis, most likely involving an earlier formation of a carotenogenic enzyme complex. Conversely, the lower carbon flux towards carotenogenesis in the deletion mutants might delay or lead to a partial formation of a carotenogenic enzyme complex, which could explain the accumulation of astaxanthin carotenoid precursors in these mutants. In conclusion, the FPS and the crtE genes represent good candidates to manipulate to favor carotenoid biosynthesis in X. dendrorhous.
Conflict of interest statement
Figures


Similar articles
-
The SREBP (Sterol Regulatory Element-Binding Protein) pathway: a regulatory bridge between carotenogenesis and sterol biosynthesis in the carotenogenic yeast Xanthophyllomyces dendrorhous.Biol Res. 2021 Oct 26;54(1):34. doi: 10.1186/s40659-021-00359-x. Biol Res. 2021. PMID: 34702374 Free PMC article. Review.
-
Engineering of geranylgeranyl pyrophosphate synthase levels and physiological conditions for enhanced carotenoid and astaxanthin synthesis in Xanthophyllomyces dendrorhous.Biotechnol Lett. 2011 Apr;33(4):755-61. doi: 10.1007/s10529-010-0495-2. Epub 2010 Dec 17. Biotechnol Lett. 2011. PMID: 21165672
-
IdsA is the major geranylgeranyl pyrophosphate synthase involved in carotenogenesis in Corynebacterium glutamicum.FEBS J. 2014 Nov;281(21):4906-20. doi: 10.1111/febs.13033. Epub 2014 Sep 30. FEBS J. 2014. PMID: 25181035
-
A Novel Hydrolytic Activity of Tri-Functional Geranylgeranyl Pyrophosphate Synthase in Haematococcus pluvialis.Plant Cell Physiol. 2018 Dec 1;59(12):2536-2548. doi: 10.1093/pcp/pcy173. Plant Cell Physiol. 2018. PMID: 30137453
-
Metabolic engineering of the astaxanthin-biosynthetic pathway of Xanthophyllomyces dendrorhous.FEMS Yeast Res. 2003 Dec;4(3):221-31. doi: 10.1016/S1567-1356(03)00158-2. FEMS Yeast Res. 2003. PMID: 14654426 Review.
Cited by
-
The Mevalonate Pathway Is Important for Growth, Spore Production, and the Virulence of Phytophthora sojae.Front Microbiol. 2021 Dec 22;12:772994. doi: 10.3389/fmicb.2021.772994. eCollection 2021. Front Microbiol. 2021. PMID: 36338274 Free PMC article.
-
The genome of the basal agaricomycete Xanthophyllomyces dendrorhous provides insights into the organization of its acetyl-CoA derived pathways and the evolution of Agaricomycotina.BMC Genomics. 2015 Mar 25;16(1):233. doi: 10.1186/s12864-015-1380-0. BMC Genomics. 2015. PMID: 25887949 Free PMC article.
-
The SREBP (Sterol Regulatory Element-Binding Protein) pathway: a regulatory bridge between carotenogenesis and sterol biosynthesis in the carotenogenic yeast Xanthophyllomyces dendrorhous.Biol Res. 2021 Oct 26;54(1):34. doi: 10.1186/s40659-021-00359-x. Biol Res. 2021. PMID: 34702374 Free PMC article. Review.
-
Morphological and Transcriptomic Analysis Reveals the Osmoadaptive Response of Endophytic Fungus Aspergillus montevidensis ZYD4 to High Salt Stress.Front Microbiol. 2017 Sep 21;8:1789. doi: 10.3389/fmicb.2017.01789. eCollection 2017. Front Microbiol. 2017. PMID: 28983284 Free PMC article.
-
The Involvement of Mig1 from Xanthophyllomyces dendrorhous in Catabolic Repression: An Active Mechanism Contributing to the Regulation of Carotenoid Production.PLoS One. 2016 Sep 13;11(9):e0162838. doi: 10.1371/journal.pone.0162838. eCollection 2016. PLoS One. 2016. PMID: 27622474 Free PMC article.
References
-
- Misawa N (2011) Pathway engineering for functional isoprenoids. Current Opinion in Biotechnology 22: 627–633. - PubMed
-
- Sacchettini JC, Poulter CD (1997) Creating isoprenoid diversity. Science 277: 1788–1789. - PubMed
-
- Liang P, Ko T, Wang AH (2002) Structure, mechanism and function of prenyltransferases. European Journal of Biochemistry 269: 3339–3354. - PubMed
-
- Lee P, Schmidt-Dannert C (2002) Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Applied Microbiology and Biotechnology 60: 1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

