Fidelity of histone gene regulation is obligatory for genome replication and stability
- PMID: 24797072
- PMCID: PMC4097655
- DOI: 10.1128/MCB.01567-13
Fidelity of histone gene regulation is obligatory for genome replication and stability
Abstract
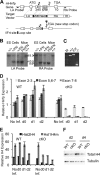
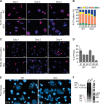
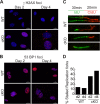
Fidelity of chromatin organization is crucial for normal cell cycle progression, and perturbations in packaging of DNA may predispose to transformation. Histone H4 protein is the most highly conserved chromatin protein, required for nucleosome assembly, with multiple histone H4 gene copies encoding identical protein. There is a long-standing recognition of the linkage of histone gene expression and DNA replication. A fundamental and unresolved question is the mechanism that couples histone biosynthesis with DNA replication and fidelity of cell cycle control. Here, we conditionally ablated the obligatory histone H4 transcription factor HINFP to cause depletion of histone H4 in mammalian cells. Deregulation of histone H4 results in catastrophic cellular and molecular defects that lead to genomic instability. Histone H4 depletion increases nucleosome spacing, impedes DNA synthesis, alters chromosome complement, and creates replicative stress. Our study provides functional evidence that the tight coupling between DNA replication and histone synthesis is reciprocal.
Figures



References
-
- Wolffe A. 1995. Chromatin: structure and function. Academic Press, London, United Kingdom
-
- Stein GS. 1984. Histone genes: structure, organization, and regulation. John Wiley & Sons, New York, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
