Synapsis alters RAG-mediated nicking at Tcrb recombination signal sequences: implications for the “beyond 12/23” rule
- PMID: 24797073
- PMCID: PMC4097660
- DOI: 10.1128/MCB.00411-14
Synapsis alters RAG-mediated nicking at Tcrb recombination signal sequences: implications for the “beyond 12/23” rule
Abstract
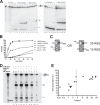
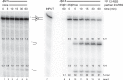
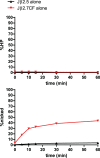
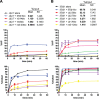
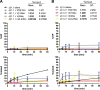
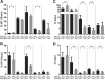
At the Tcrb locus, Vβ-to-Jβ rearrangement is permitted by the 12/23 rule but is not observed in vivo, a restriction termed the “beyond 12/23” rule (B12/23 rule). Previous work showed that Vβ recombination signal sequences (RSSs) do not recombine with Jβ RSSs because Jβ RSSs are crippled for either nicking or synapsis. This result raised the following question: how can crippled Jβ RSSs recombine with Dβ RSSs? We report here that the nicking of some Jβ RSSs can be substantially stimulated by synapsis with a 3′Dβ1 partner RSS. This result helps to reconcile disagreement in the field regarding the impact of synapsis on nicking. Furthermore, our data allow for the classification of Tcrb RSSs into two major categories: those that nick quickly and those that nick slowly in the absence of a partner. Slow-nicking RSSs can be stimulated to nick more efficiently upon synapsis with an appropriate B12/23 partner, and our data unexpectedly suggest that fast-nicking RSSs can be inhibited for nicking upon synapsis with an inappropriate partner. These observations indicate that the RAG proteins exert fine control over every step of V(D)J cleavage and support the hypothesis that initial RAG binding can occur on RSSs with either 12- or 23-bp spacers (12- or 23-RSSs, respectively).
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
