The intraviral protein interaction network of hepatitis C virus
- PMID: 24797426
- PMCID: PMC4083108
- DOI: 10.1074/mcp.M113.036301
The intraviral protein interaction network of hepatitis C virus
Abstract
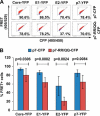
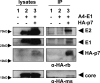
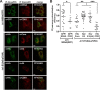
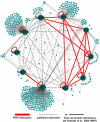
Hepatitis C virus (HCV) is a global health problem and one of the main reasons for chronic liver diseases such as cirrhosis and hepatocellular carcinoma. The HCV genome is translated into a polyprotein which is proteolytically processed into 10 viral proteins. The interactome of the HCV proteins with the host cell has been worked out; however, it remains unclear how viral proteins interact with each other. We aimed to generate the interaction network of these 10 HCV proteins using a flow-cytometry-based FRET assay established in our laboratory (Banning, C., Votteler, J., Hoffmann, D., Koppensteiner, H., Warmer, M., Reimer, R., Kirchhoff, F., Schubert, U., Hauber, J., and Schindler, M. (2010) A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5, e9344). HCV proteins were constructed as fusions with the chromophores CFP and YFP. All HCV fusions were expressed and localized to specific subcellular compartments, indicating that they were functional. FACS-FRET measurements identified a total of 20 interactions; 13 of these were previously described and have now been confirmed in living cells via our method. Among the seven novel protein binding pairs, HCV p7 plays a pivotal role. It binds to the HCV capsid protein Core and the two glycoproteins E1 and E2. These interplays were further demonstrated in the relevant context of Huh7.5 liver cells expressing infectious HCV. Our work demonstrates the feasibility of rapidly generating small interaction networks via FACS-FRET and defines the network of intra-HCV protein interactions. Furthermore, our data support an important role of p7 in HCV assembly.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Construction and analysis of a comprehensive protein interaction network of HCV with its host Homo sapiens.BMC Infect Dis. 2019 Apr 30;19(1):367. doi: 10.1186/s12879-019-4000-9. BMC Infect Dis. 2019. PMID: 31039741 Free PMC article.
-
Detergent-resistant membrane association of NS2 and E2 during hepatitis C virus replication.J Virol. 2015 Apr;89(8):4562-74. doi: 10.1128/JVI.00123-15. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673706 Free PMC article.
-
Diffusion of information throughout the host interactome reveals gene expression variations in network proximity to target proteins of hepatitis C virus.PLoS One. 2014 Dec 2;9(12):e113660. doi: 10.1371/journal.pone.0113660. eCollection 2014. PLoS One. 2014. PMID: 25461596 Free PMC article.
-
Molecular virology of hepatitis C virus.Acta Med Okayama. 2001 Jun;55(3):133-59. doi: 10.18926/AMO/32025. Acta Med Okayama. 2001. PMID: 11434427 Review.
-
Post-translational modifications of hepatitis C viral proteins and their biological significance.World J Gastroenterol. 2013 Dec 21;19(47):8929-39. doi: 10.3748/wjg.v19.i47.8929. World J Gastroenterol. 2013. PMID: 24379618 Free PMC article. Review.
Cited by
-
The Tetherin Antagonism of the Ebola Virus Glycoprotein Requires an Intact Receptor-Binding Domain and Can Be Blocked by GP1-Specific Antibodies.J Virol. 2016 Nov 28;90(24):11075-11086. doi: 10.1128/JVI.01563-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707924 Free PMC article.
-
Beyond Channel Activity: Protein-Protein Interactions Involving Viroporins.Subcell Biochem. 2018;88:329-377. doi: 10.1007/978-981-10-8456-0_15. Subcell Biochem. 2018. PMID: 29900504 Free PMC article. Review.
-
Hepatitis C Virus Is Released via a Noncanonical Secretory Route.J Virol. 2016 Nov 14;90(23):10558-10573. doi: 10.1128/JVI.01615-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27630244 Free PMC article.
-
Coevolution analysis of Hepatitis C virus genome to identify the structural and functional dependency network of viral proteins.Sci Rep. 2016 May 20;6:26401. doi: 10.1038/srep26401. Sci Rep. 2016. PMID: 27198619 Free PMC article.
-
Combinatorial interactions between viral proteins expand the potential functional landscape of the tomato yellow leaf curl virus proteome.PLoS Pathog. 2022 Oct 18;18(10):e1010909. doi: 10.1371/journal.ppat.1010909. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36256684 Free PMC article.
References
-
- Moradpour D., Penin F., Rice C. M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 - PubMed
-
- Kronenberger B., Zeuzem S. (2012) New developments in HCV therapy. J. Viral Hepatitis 19 Suppl 1, 48–51 - PubMed
-
- de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugue S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., Andre P., Rabourdin-Combe C., Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

