The silicon trypanosome: a test case of iterative model extension in systems biology
- PMID: 24797926
- PMCID: PMC4773886
- DOI: 10.1016/B978-0-12-800143-1.00003-8
The silicon trypanosome: a test case of iterative model extension in systems biology
Abstract
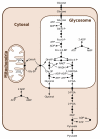
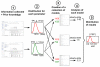
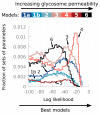
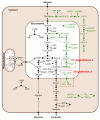
The African trypanosome, Trypanosoma brucei, is a unicellular parasite causing African Trypanosomiasis (sleeping sickness in humans and nagana in animals). Due to some of its unique properties, it has emerged as a popular model organism in systems biology. A predictive quantitative model of glycolysis in the bloodstream form of the parasite has been constructed and updated several times. The Silicon Trypanosome is a project that brings together modellers and experimentalists to improve and extend this core model with new pathways and additional levels of regulation. These new extensions and analyses use computational methods that explicitly take different levels of uncertainty into account. During this project, numerous tools and techniques have been developed for this purpose, which can now be used for a wide range of different studies in systems biology.
Keywords: Differential equations; Dynamic modelling; Metabolomics; Systems biology; Transcriptomics; Trypanosoma brucei; Uncertainty.
© 2014 Elsevier Ltd All rights reserved.
Figures


References
-
- Barrett MP, Burchmore RJS, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. Lancet. 2003 Nov;362(9394):1469–1480. - PubMed
-
- Barrett MP, Bakker BM, Breitling R. Metabolomic systems biology of trypanosomes. Parasitology. 2010 Aug;137(9):1285–1290. - PubMed
-
- Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. Glycolysis in bloodstream form Trypanosoma brucei can be understood in terms of the kinetics of the glycolytic enzymes. J. Biol. Chem. 1997 Feb;272(6):3207–3215. - PubMed
-
- Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. What controls glycolysis in bloodstream form Trypanosoma brucei? J Biol Chem. 1999 May;274(21):14551–14559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

