Structure and function of preQ1 riboswitches
- PMID: 24798077
- PMCID: PMC4177978
- DOI: 10.1016/j.bbagrm.2014.04.019
Structure and function of preQ1 riboswitches
Abstract
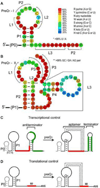
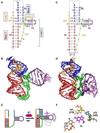
PreQ1 riboswitches help regulate the biosynthesis and transport of preQ1 (7-aminomethyl-7-deazaguanine), a precursor of the hypermodified guanine nucleotide queuosine (Q), in a number of Firmicutes, Proteobacteria, and Fusobacteria. Queuosine is almost universally found at the wobble position of the anticodon in asparaginyl, tyrosyl, histidyl and aspartyl tRNAs, where it contributes to translational fidelity. Two classes of preQ1 riboswitches have been identified (preQ1-I and preQ1-II), and structures of examples from both classes have been determined. Both classes form H-type pseudoknots upon preQ1 binding, each of which has distinct unusual features and modes of preQ1 recognition. These features include an unusually long loop 2 in preQ1-I pseudoknots and an embedded hairpin in loop 3 in preQ1-II pseudoknots. PreQ1-I riboswitches are also notable for their unusually small aptamer domain, which has been extensively investigated by NMR, X-ray crystallography, FRET, and other biophysical methods. Here we review the discovery, structural biology, ligand specificity, cation interactions, folding, dynamics, and applications to biotechnology of preQ1 riboswitches. This article is part of a Special Issue entitled: Riboswitches.
Keywords: NMR; PreQ(0); Queuine; Queuosine; X-ray crystallography; tRNA modification.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures



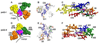

References
-
- Kim JN, Breaker RR. Purine sensing by riboswitches. Biology of the cell / under the auspices of the European Cell Biology Organization. 2008;100:1–11. - PubMed
-
- Harada F, Nishimura S. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry. 1972;11:301–308. - PubMed
-
- Morris RC, Elliott MS. Queuosine modification of tRNA: a case for convergent evolution. Mol Genet Metab. 2001;74:147–159. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

