Simulations of remote mutants of dihydrofolate reductase reveal the nature of a network of residues coupled to hydride transfer
- PMID: 24798860
- PMCID: PMC4082691
- DOI: 10.1002/jcc.23629
Simulations of remote mutants of dihydrofolate reductase reveal the nature of a network of residues coupled to hydride transfer
Abstract
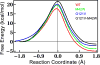
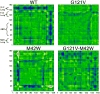
Recent experimental and theoretical studies have proposed that enzymes involve networks of coupled residues throughout the protein that participate in motions accompanying chemical barrier crossing. Here, we have examined portions of a proposed network in dihydrofolate reductase (DHFR) using quantum mechanics/molecular mechanics simulations. The simulations use a hybrid quantum mechanics-molecular mechanics approach with a recently developed semiempirical AM1-SRP Hamiltonian that provides accurate results for this reaction. The simulations reproduce experimentally determined catalytic rates for the wild type and distant mutants of E. coli DHFR, underscoring the accuracy of the simulation protocol. Additionally, the simulations provide detailed insight into how residues remote from the active site affect the catalyzed chemistry, through changes in the thermally averaged properties along the reaction coordinate. The mutations do not greatly affect the structure of the transition state near the bond activation, but we observe differences somewhat removed from the point of C-H cleavage that affect the rate. The mutations have global effects on the thermally averaged structure that propagate throughout the enzyme and the current simulations highlight several interactions that appear to be particularly important.
Keywords: AM1-SRP; QM/MM; enzyme dynamics; hydrogen tunneling; network of interactions.
Copyright © 2014 Wiley Periodicals, Inc.
Figures

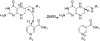
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

