The additive damage model: a mathematical model for cellular responses to drug combinations
- PMID: 24799130
- PMCID: PMC4367491
- DOI: 10.1016/j.jtbi.2014.04.032
The additive damage model: a mathematical model for cellular responses to drug combinations
Abstract
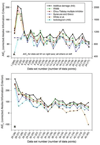
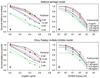
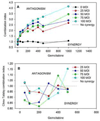
Mathematical models to describe dose-dependent cellular responses to drug combinations are an essential component of computational simulations for predicting therapeutic responses. Here, a new model, the additive damage model, is introduced and tested in cases where varying concentrations of two drugs are applied with a fixed exposure schedule. In the model, cell survival is determined by whether cellular damage, which depends on the concentrations of the drugs, exceeds a lethal threshold, which varies randomly in the cell population with a prescribed statistical distribution. Cellular damage is assumed to be additive, and is expressed as a sum of separate terms for each drug. Each term has a saturable dependence on drug concentration. The model has appropriate behavior over the entire range of drug concentrations, and is predictive, given single-agent dose-response data for each drug. The proposed model is compared with several other models, by testing their ability to fit 24 data sets for platinum-taxane combinations and 21 data sets for various other combinations. The Akaike Information Criterion is used to assess goodness of fit, taking into account the number of unknown parameters in each model. Overall, the additive damage model provides a better fit to the data sets than any previous model. The proposed model provides a basis for computational simulations of therapeutic responses. It predicts responses to drug combinations based on data for each drug acting as a single agent, and can be used as an improved null reference model for assessing synergy in the action of drug combinations.
Keywords: Cellular pharmacodynamics; Chemotherapy; Cytotoxicity; Dose–response; Drug synergy.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Additive Damage Models for Cellular Pharmacodynamics of Radiation-Chemotherapy Combinations.Bull Math Biol. 2018 May;80(5):1236-1258. doi: 10.1007/s11538-017-0316-z. Epub 2017 Aug 28. Bull Math Biol. 2018. PMID: 28849417
-
Two-mechanism peak concentration model for cellular pharmacodynamics of Doxorubicin.Neoplasia. 2005 Jul;7(7):705-13. doi: 10.1593/neo.05118. Neoplasia. 2005. PMID: 16026650 Free PMC article.
-
Cell cycle checkpoint models for cellular pharmacology of paclitaxel and platinum drugs.AAPS J. 2008;10(1):15-34. doi: 10.1208/s12248-007-9003-6. Epub 2008 Feb 5. AAPS J. 2008. PMID: 18446502 Free PMC article.
-
Advances in computational approaches in identifying synergistic drug combinations.Brief Bioinform. 2018 Nov 27;19(6):1172-1182. doi: 10.1093/bib/bbx047. Brief Bioinform. 2018. PMID: 28475767 Review.
-
Taxane-platinum combinations in advanced non-small cell lung cancer: a review.Oncologist. 2004;9 Suppl 2:16-23. doi: 10.1634/theoncologist.9-suppl_2-16. Oncologist. 2004. PMID: 15161987 Review.
Cited by
-
Experimentally-driven mathematical modeling to improve combination targeted and cytotoxic therapy for HER2+ breast cancer.Sci Rep. 2019 Sep 6;9(1):12830. doi: 10.1038/s41598-019-49073-5. Sci Rep. 2019. PMID: 31492947 Free PMC article.
-
Tissue transport affects how treatment scheduling increases the efficacy of chemotherapeutic drugs.J Theor Biol. 2018 Feb 7;438:21-33. doi: 10.1016/j.jtbi.2017.10.022. Epub 2017 Oct 21. J Theor Biol. 2018. PMID: 29066114 Free PMC article.
-
Leveraging Mathematical Modeling to Quantify Pharmacokinetic and Pharmacodynamic Pathways: Equivalent Dose Metric.Front Physiol. 2019 May 22;10:616. doi: 10.3389/fphys.2019.00616. eCollection 2019. Front Physiol. 2019. PMID: 31178753 Free PMC article.
-
Advances in Synergistic Combinations of Chinese Herbal Medicine for the Treatment of Cancer.Curr Cancer Drug Targets. 2016;16(4):346-56. doi: 10.2174/1568009616666151207105851. Curr Cancer Drug Targets. 2016. PMID: 26638885 Free PMC article. Review.
References
-
- White DB, Slocum HK, Brun Y, Wrzosek C, Greco WR. A new nonlinear mixture response surface paradigm for the study of synergism: A three drug example. Current Drug Metabolism. 2003;4:399–409. - PubMed
-
- Chou TC, Talalay P. Generalized Equations for the Analysis of Inhibitions of Michaelis-Menten and Higher-Order Kinetic Systems with 2 Or More Mutually Exclusive and Non-Exclusive Inhibitors. European Journal of Biochemistry. 1981;115:207–216. - PubMed
-
- Greco WR, Faessel H, Levasseur L. The search for cytotoxic synergy between anticancer agents: A case of Dorothy and the ruby slippers? Journal of the National Cancer Institute. 1996;88:699–700. - PubMed
-
- Webb JL. Effect of more than one inhibitor. Vol. 66. New York: Academic Press; 1963. p. 487.
-
- Greco WR, Park HS, Rustum YM. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1990;50:5318–5327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

