A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development
- PMID: 24799598
- PMCID: PMC6083926
- DOI: 10.1093/infdis/jiu260
A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development
Abstract
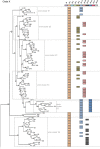
Streptococcus pyogenes ranks among the main causes of mortality from bacterial infections worldwide. Currently there is no vaccine to prevent diseases such as rheumatic heart disease and invasive streptococcal infection. The streptococcal M protein that is used as the substrate for epidemiological typing is both a virulence factor and a vaccine antigen. Over 220 variants of this protein have been described, making comparisons between proteins difficult, and hindering M protein-based vaccine development. A functional classification based on 48 emm-clusters containing closely related M proteins that share binding and structural properties is proposed. The need for a paradigm shift from type-specific immunity against S. pyogenes to emm-cluster based immunity for this bacterium should be further investigated. Implementation of this emm-cluster-based system as a standard typing scheme for S. pyogenes will facilitate the design of future studies of M protein function, streptococcal virulence, epidemiological surveillance, and vaccine development.
Keywords: IgA; IgG; M protein; Streptococcus pyogenes; epidemiology; fibrinogen; molecular typing; plasminogen; vaccine.
© The Author 2014. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures







References
-
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. - PubMed
-
- Dale JB, Fischetti VA, Carapetis JR, et al. Group A streptococcal vaccines: paving a path for accelerated development. Vaccine. 2013;31(suppl 2):B216–22. - PubMed
-
- Smeesters PR, McMillan DJ, Sriprakash KS. The streptococcal M protein: a highly versatile molecule. Trends Microbiol. 2010;18:275–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

