Profiling of olfactory receptor gene expression in whole human olfactory mucosa
- PMID: 24800820
- PMCID: PMC4011832
- DOI: 10.1371/journal.pone.0096333
Profiling of olfactory receptor gene expression in whole human olfactory mucosa
Abstract
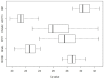
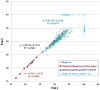
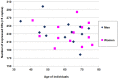
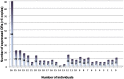
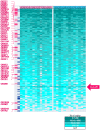
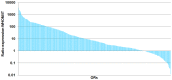
Olfactory perception is mediated by a large array of olfactory receptor genes. The human genome contains 851 olfactory receptor gene loci. More than 50% of the loci are annotated as nonfunctional due to frame-disrupting mutations. Furthermore haplotypic missense alleles can be nonfunctional resulting from substitution of key amino acids governing protein folding or interactions with signal transduction components. Beyond their role in odor recognition, functional olfactory receptors are also required for a proper targeting of olfactory neuron axons to their corresponding glomeruli in the olfactory bulb. Therefore, we anticipate that profiling of olfactory receptor gene expression in whole human olfactory mucosa and analysis in the human population of their expression should provide an opportunity to select the frequently expressed and potentially functional olfactory receptors in view of a systematic deorphanization. To address this issue, we designed a TaqMan Low Density Array (Applied Biosystems), containing probes for 356 predicted human olfactory receptor loci to investigate their expression in whole human olfactory mucosa tissues from 26 individuals (13 women, 13 men; aged from 39 to 81 years, with an average of 67±11 years for women and 63±12 years for men). Total RNA isolation, DNase treatment, RNA integrity evaluation and reverse transcription were performed for these 26 samples. Then 384 targeted genes (including endogenous control genes and reference genes specifically expressed in olfactory epithelium for normalization purpose) were analyzed using the same real-time reverse transcription PCR platform. On average, the expression of 273 human olfactory receptor genes was observed in the 26 selected whole human olfactory mucosa analyzed, of which 90 were expressed in all 26 individuals. Most of the olfactory receptors deorphanized to date on the basis of sensitivity to known odorant molecules, which are described in the literature, were found in the expressed olfactory receptors gene set.
Conflict of interest statement
Figures

Similar articles
-
Olfactory discrimination largely persists in mice with defects in odorant receptor expression and axon guidance.Neural Dev. 2012 Jul 4;7:17. doi: 10.1186/1749-8104-7-17. Neural Dev. 2012. PMID: 22559903 Free PMC article.
-
The human olfactory transcriptome.BMC Genomics. 2016 Aug 11;17(1):619. doi: 10.1186/s12864-016-2960-3. BMC Genomics. 2016. PMID: 27515280 Free PMC article.
-
Coordination of olfactory receptor choice with guidance receptor expression and function in olfactory sensory neurons.PLoS Genet. 2018 Jan 31;14(1):e1007164. doi: 10.1371/journal.pgen.1007164. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29385124 Free PMC article.
-
Olfactory receptors: molecular basis for recognition and discrimination of odors.Anal Bioanal Chem. 2003 Oct;377(3):427-33. doi: 10.1007/s00216-003-2113-9. Epub 2003 Aug 1. Anal Bioanal Chem. 2003. PMID: 12898108 Review.
-
Olfactory receptor function.Handb Clin Neurol. 2019;164:67-78. doi: 10.1016/B978-0-444-63855-7.00005-8. Handb Clin Neurol. 2019. PMID: 31604564 Review.
Cited by
-
Receptor arrays optimized for natural odor statistics.Proc Natl Acad Sci U S A. 2016 May 17;113(20):5570-5. doi: 10.1073/pnas.1600357113. Epub 2016 Apr 21. Proc Natl Acad Sci U S A. 2016. PMID: 27102871 Free PMC article.
-
Olfaction in (Social) Context: The Role of Social Complexity in Trajectories of Older Adults' Olfactory Abilities.J Aging Health. 2023 Jan;35(1-2):108-124. doi: 10.1177/08982643221108020. Epub 2022 Jun 23. J Aging Health. 2023. PMID: 35739641 Free PMC article.
-
Normalized Neural Representations of Complex Odors.PLoS One. 2016 Nov 11;11(11):e0166456. doi: 10.1371/journal.pone.0166456. eCollection 2016. PLoS One. 2016. PMID: 27835696 Free PMC article.
-
Olfactory Function and Olfactory Disorders.Laryngorhinootologie. 2023 May;102(S 01):S67-S92. doi: 10.1055/a-1957-3267. Epub 2023 May 2. Laryngorhinootologie. 2023. PMID: 37130532 Free PMC article.
-
Correlation between olfactory function, age, sex, and cognitive reserve index in the Italian population.Eur Arch Otorhinolaryngol. 2022 Oct;279(10):4943-4952. doi: 10.1007/s00405-022-07311-z. Epub 2022 Feb 24. Eur Arch Otorhinolaryngol. 2022. PMID: 35211821 Free PMC article.
References
-
- Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175–187. - PubMed
-
- Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, et al. (1994) Olfactory receptor gene cluster on human chromosome 17: possible duplication of an ancestral receptor repertoire. Hum Mol Genet 3: 229–235. - PubMed
-
- Glusman G, Bahar A, Sharon D, Pilpel Y, White J, et al. (2000) The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome 11: 1016–1023. - PubMed
-
- Rouquier S, Taviaux S, Trask BJ, Brand-Arpon V, van den Engh G, et al. (1998) Distribution of olfactory receptor genes in the human genome. Nat Genet 18: 243–250. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

