Chronic lymphocytic leukemia cells in a lymph node microenvironment depict molecular signature associated with an aggressive disease
- PMID: 24800836
- PMCID: PMC4107103
- DOI: 10.2119/molmed.2012.00303
Chronic lymphocytic leukemia cells in a lymph node microenvironment depict molecular signature associated with an aggressive disease
Abstract
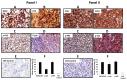
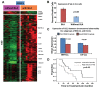
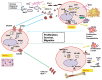
Chronic lymphocytic leukemia (CLL) cells survive longer in vivo than in vitro, suggesting that the tissue microenvironment provides prosurvival signals to tumor cells. Primary and secondary lymphoid tissues are involved in the pathogenesis of CLL, and the role of these tissue microenvironments has not been explored completely. To elucidate host-tumor interactions, we performed gene expression profiling (GEP) of purified CLL cells from peripheral blood (PB; n = 20), bone marrow (BM; n = 18), and lymph node (LN; n = 15) and validated key pathway genes by real-time polymerase chain reaction, immunohistochemistry and/or TCL1 trans-genic mice. Gene signatures representing several pathways critical for survival and activation of B cells were altered in CLL cells from different tissue compartments. Molecules associated with the B-cell receptor (BCR), B cell-activating factor/a proliferation-inducing ligand (BAFF/APRIL), nuclear factor (NF)-κB pathway and immune suppression signature were enriched in LN-CLL, suggesting LNs as the primary site for tumor growth. Immune suppression genes may help LN-CLL cells to modulate antigen-presenting and T-cell behavior to suppress antitumor activity. PB CLL cells overexpressed chemokine receptors, and their cognate ligands were enriched in LN and BM, suggesting that a chemokine gradient instructs B cells to migrate toward LN or BM. Of several chemokine ligands, the expression of CCL3 was associated with poor prognostic factors. The BM gene signature was enriched with antiapoptotic, cytoskeleton and adhesion molecules. Interestingly, PB cells from lymphadenopathy patients shared GEP with LN cells. In Eμ-TCL1 transgenic mice (the mouse model of the disease), a high percentage of leukemic cells from the lymphoid compartment express key BCR and NF-κB molecules. Together, our findings demonstrate that the lymphoid microenvironment promotes survival, proliferation and progression of CLL cells via chronic activation of BCR, BAFF/APRIL and NF-κB activation while suppressing the immune response.
Figures



References
-
- Chiorazzi N. Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best Pract Res Clin Haematol. 2007;20:399–413. - PubMed
-
- Defoiche J, et al. Reduction of B cell turnover in chronic lymphocytic leukaemia. Br J Haematol. 2008;143:240–7. - PubMed
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. - PubMed
-
- Burger JA, Kipps TJ. Chemokine receptors and stromal cells in the homing and homeostasis of chronic lymphocytic leukemia B cells. Leuk Lymphoma. 2002;43:461–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

