Differential expression profiling of spleen microRNAs in response to two distinct type II interferons in Tetraodon nigroviridis
- PMID: 24800866
- PMCID: PMC4011704
- DOI: 10.1371/journal.pone.0096336
Differential expression profiling of spleen microRNAs in response to two distinct type II interferons in Tetraodon nigroviridis
Abstract
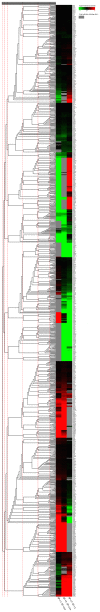
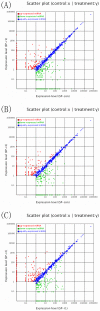
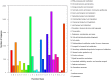
MicroRNAs are endogenous, small non-coding RNAs approximately 18-26 nucleotides in length that regulate target gene expression at the post-transcription level. Interferon-γ (IFN-γ) is a Th1 cytokine that is involved in both the innate and adaptive immune responses. We previously identified two IFN-γ genes in green-spotted puffer fish (Tetraodon nigroviridis). To determine whether miRNAs participate in IFN-γ-related immune responses, T. nigroviridis spleen cells were treated with recombinant IFN-γ isoforms, and a Solexa high-throughput sequencing method was used to identify miRNAs. In total, 1,556, 1,538 and 1,573 miRNAs were found in the three samples, and differentially expressed miRNAs were determined. In total, 398 miRNAs were differentially expressed after rIFN-γ1 treatment, and 438 miRNAs were differentially expressed after rIFN-γ2 treatment; additionally, 403 miRNAs were differentially expressed between the treatment groups. Ten differentially expressed miRNAs were chosen for validation using qRT-PCR. Target genes for the differentially expressed miRNAs were predicted, and GO and KEGG analyses were performed. This study provides basic knowledge regarding fish IFN-γ-induced miRNAs and offers clues for further studies into the mechanisms underlying fish IFN-γ-mediated immune responses.
Conflict of interest statement
Figures




Similar articles
-
Two distinct interferon-γ genes in Tetraodon nigroviridis: Functional analysis during Vibrio parahaemolyticus infection.Mol Immunol. 2016 Feb;70:34-46. doi: 10.1016/j.molimm.2015.12.004. Epub 2015 Dec 14. Mol Immunol. 2016. PMID: 26701668
-
Identification and characterization of microRNAs in the spleen of common carp immune organ.J Cell Biochem. 2014 Oct;115(10):1768-78. doi: 10.1002/jcb.24843. J Cell Biochem. 2014. PMID: 24819892
-
Transcriptome-Wide Comparative Analysis of microRNA Profiles in the Telogen Skins of Liaoning Cashmere Goats (Capra hircus) and Fine-Wool Sheep (Ovis aries) by Solexa Deep Sequencing.DNA Cell Biol. 2016 Nov;35(11):696-705. doi: 10.1089/dna.2015.3161. Epub 2016 Oct 18. DNA Cell Biol. 2016. PMID: 27754706
-
Differential regulation of Tetraodon nigroviridis Mx gene promoter activity by constitutively-active forms of STAT1, STAT2, and IRF9.Fish Shellfish Immunol. 2014 May;38(1):230-43. doi: 10.1016/j.fsi.2014.03.017. Epub 2014 Mar 27. Fish Shellfish Immunol. 2014. PMID: 24680831
-
Gill transcriptome response to changes in environmental calcium in the green spotted puffer fish.BMC Genomics. 2010 Aug 17;11:476. doi: 10.1186/1471-2164-11-476. BMC Genomics. 2010. PMID: 20716350 Free PMC article.
Cited by
-
Expression characteristics and interaction networks of microRNAs in spleen tissues of grass carp (Ctenopharyngodon idella).PLoS One. 2022 Mar 28;17(3):e0266189. doi: 10.1371/journal.pone.0266189. eCollection 2022. PLoS One. 2022. PMID: 35344574 Free PMC article.
-
Two Distinct Interferon-γ in the Orange-Spotted Grouper (Epinephelus coioides): Molecular Cloning, Functional Characterization, and Regulation in Toll-Like Receptor Pathway by Induction of miR-146a.Front Endocrinol (Lausanne). 2018 Feb 26;9:41. doi: 10.3389/fendo.2018.00041. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 29535680 Free PMC article.
-
High-Throughput Identification of MiR-145 Targets in Human Articular Chondrocytes.Life (Basel). 2020 May 11;10(5):58. doi: 10.3390/life10050058. Life (Basel). 2020. PMID: 32403239 Free PMC article.
References
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
-
- Banerjee D, Kwok A, Lin SY, Slack FJ (2005) Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell 8: 287–295. - PubMed
-
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338. - PubMed
-
- Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86. - PubMed
-
- Lee YS, Kim HK, Chung S, Kim KS, Dutta A (2005) Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280: 16635–16641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

