Site-specific promoter caging enables optochemical gene activation in cells and animals
- PMID: 24802207
- PMCID: PMC4333597
- DOI: 10.1021/ja500327g
Site-specific promoter caging enables optochemical gene activation in cells and animals
Abstract
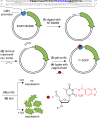
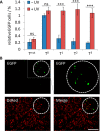
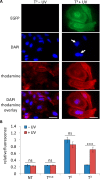
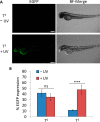
In cell and molecular biology, double-stranded circular DNA constructs, known as plasmids, are extensively used to express a gene of interest. These gene expression systems rely on a specific promoter region to drive the transcription of genes either constitutively (i.e., in a continually "ON" state) or conditionally (i.e., in response to a specific transcription initiator). However, controlling plasmid-based expression with high spatial and temporal resolution in cellular environments and in multicellular organisms remains challenging. To overcome this limitation, we have site-specifically installed nucleobase-caging groups within a plasmid promoter region to enable optochemical control of transcription and, thus, gene expression, via photolysis of the caging groups. Through the light-responsive modification of plasmid-based gene expression systems, we have demonstrated optochemical activation of an exogenous fluorescent reporter gene in both tissue culture and a live animal model, as well as light-induced overexpression of an endogenous signaling protein.
Figures
Similar articles
-
Construction of a new minicircle DNA carrying an enhanced green florescent protein reporter gene for efficient expression into mammalian cell lines.Mol Biol Rep. 2015 Jul;42(7):1175-85. doi: 10.1007/s11033-015-3864-z. Epub 2015 Mar 4. Mol Biol Rep. 2015. PMID: 25736052
-
Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach.Acc Chem Res. 2014 Jan 21;47(1):45-55. doi: 10.1021/ar400036a. Epub 2013 Aug 28. Acc Chem Res. 2014. PMID: 23981235 Free PMC article.
-
cis-Acting Determinant Limiting Expression of Sphingomyelinase Gene sph2 in Leptospira interrogans, Identified with a gfp Reporter Plasmid.Appl Environ Microbiol. 2018 Nov 15;84(23):e02068-18. doi: 10.1128/AEM.02068-18. Print 2018 Dec 1. Appl Environ Microbiol. 2018. PMID: 30266732 Free PMC article.
-
Optochemical control of RNA interference in mammalian cells.Nucleic Acids Res. 2013 Dec;41(22):10518-28. doi: 10.1093/nar/gkt806. Epub 2013 Sep 10. Nucleic Acids Res. 2013. PMID: 24021631 Free PMC article.
-
Coumarin-Derived Caging Groups in the Spotlight: Tailoring Physiochemical and Photophysical Properties.Chempluschem. 2024 Oct;89(10):e202400377. doi: 10.1002/cplu.202400377. Epub 2024 Aug 21. Chempluschem. 2024. PMID: 38960871 Review.
Cited by
-
Light-Activated Gene Expression System Using a Caging-Group-Free Photoactivatable Dye.Angew Chem Int Ed Engl. 2025 Jan 21;64(4):e202416420. doi: 10.1002/anie.202416420. Epub 2024 Nov 16. Angew Chem Int Ed Engl. 2025. PMID: 39444190 Free PMC article.
-
Analogs of S-Adenosyl-L-Methionine in Studies of Methyltransferases.Mol Biol. 2022;56(2):229-250. doi: 10.1134/S002689332202011X. Epub 2022 Apr 14. Mol Biol. 2022. PMID: 35440827 Free PMC article.
-
Photocleavable Anionic Glues for Light-Responsive Nanoparticle Aggregates.J Am Chem Soc. 2023 Feb 9;145(7):4098-108. doi: 10.1021/jacs.2c11973. Online ahead of print. J Am Chem Soc. 2023. PMID: 36757850 Free PMC article.
-
Controlling gene expression with light: a multidisciplinary endeavour.Biochem Soc Trans. 2020 Aug 28;48(4):1645-1659. doi: 10.1042/BST20200014. Biochem Soc Trans. 2020. PMID: 32657338 Free PMC article. Review.
-
Conditional gene knockdowns in sea urchins using caged morpholinos.Dev Biol. 2021 Jul;475:21-29. doi: 10.1016/j.ydbio.2021.02.014. Epub 2021 Mar 5. Dev Biol. 2021. PMID: 33684434 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

